Gene
KWMTBOMO05667
Pre Gene Modal
BGIBMGA006797
Annotation
PREDICTED:_tRNA-dihydrouridine(16/17)_synthase_[NAD(P)(+)]-like_isoform_X1_[Amyelois_transitella]
Full name
Palmitoyltransferase
Location in the cell
Nuclear Reliability : 3.004
Sequence
CDS
ATGGCTAACTGGATTTTTGAGTTAAATAATCCACAATTTGTTGTGGCGCCTATGGTTGATGCTAGCGAACTAGCTTGGCGCCTACTCTGTAGACGGCATGGTGCAAATTTGTGCTACACTCCTATGTTACATAGTAATGTTTTTACAAAAGATGCTAAATACAGGAAAGACAACCTAGTTACTTGTGAAGAAGATCGTCCTTTAATAGTACAGTTTTGCGGTAACAACCCTGAAATTATGGCATCAGCTGCAAAATTAGCTGAAGAATATTGTGATGCTATAGACATCAACCTTGGATGTCCTCAATCTATTGCTAAAAGAGGAAGATATGGATCCTTTCTTCAAGATGACTGGGAATTATTGAAAGAGATCGTGACTGCAATGTCCAAGGCTGTTTCCATACCAATCACATGTAAAGTGAGAATATTTGAAGATATTGACAAATCAGTAAATTATGCCAAGATGCTAGAAGCATCTGGATGTAAACTCCTAACAGTTCATGGAAGAACAAGAGAGCAGAAAGGGCCTTTGACTGGCATTGCTAGTTGGAAACATATAAAAGCTATAAGGGAGTCTGTATCAATACCTATGCTTGCAAATGGTAATATTCAGTGTCTCGAAGATGTATACAGGTGTATAGAATATACCAAAGTTGATGGCGTAATGAGTGCAGAGGGAATTCTTACAAATCCTGCTCTGTTTGAAGGTATTAACCCAGTCACTTGGGAATTGGCAGAAGAGTATTTAGAATTAGTTTACAGGTATCCCTGCCCCTCATCTTATGTTAGAGGACATCTATTCAAAATTTTCCATAAAATTTTCACCTTTGATGAGAACAATGATGACCGACAATTACTTGCTACAGCTCAGAACATAAATGACTTCAAGAAAGTTTGTGAAAATCTAAAAAATAAATATCAACCTTATCAAGAAGGGAAAATACATTATGAAGATGAAGAAAATATATCCAGGACAGGGTACAATTTAATTTTACCACCATGGATATGTCAACCTTATGTGAGAATGTCACCAGAAGCACATACACAGAAAATGCACGAGCTAAGTAAAAATCAGAACAATCAGGAATGTAAAAGAATTAATGAAGACCAAGATGGAAATCATATATCACGTAAGAAAATGAAAAAGATGAAACGGATGATGCGAAGACCTCAGAAAGCAGATCAAAACCAGAATGGGCGAAATGGAGATATTTGTTTCAATGATAAATGTCCTAATCCTCTGGGCGGTAAATGTACATACAAATTATGTAAAAAATGTTGTAAAAATAAGTGTTACGAAGAAGACTTGGATTGTAGAGGTCACAGGATACTTGTTCATACTAGAAGAGAAATTGCCAAATTATTTGCAAATAAAAATACTGAGGAGACGTAG
Protein
MANWIFELNNPQFVVAPMVDASELAWRLLCRRHGANLCYTPMLHSNVFTKDAKYRKDNLVTCEEDRPLIVQFCGNNPEIMASAAKLAEEYCDAIDINLGCPQSIAKRGRYGSFLQDDWELLKEIVTAMSKAVSIPITCKVRIFEDIDKSVNYAKMLEASGCKLLTVHGRTREQKGPLTGIASWKHIKAIRESVSIPMLANGNIQCLEDVYRCIEYTKVDGVMSAEGILTNPALFEGINPVTWELAEEYLELVYRYPCPSSYVRGHLFKIFHKIFTFDENNDDRQLLATAQNINDFKKVCENLKNKYQPYQEGKIHYEDEENISRTGYNLILPPWICQPYVRMSPEAHTQKMHELSKNQNNQECKRINEDQDGNHISRKKMKKMKRMMRRPQKADQNQNGRNGDICFNDKCPNPLGGKCTYKLCKKCCKNKCYEEDLDCRGHRILVHTRREIAKLFANKNTEET
Summary
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Similarity
Belongs to the DHHC palmitoyltransferase family.
Uniprot
H9JBA2
A0A2H1VAJ1
A0A2A4JMB6
A0A3S2LSS1
S4NTX3
A0A212ERP2
+ More
A0A194QL60 A0A194PKA3 A0A2A4JNY5 A0A0L7KQ81 D6WDX9 A0A026WXE5 B4N791 A0A0A9Y6K3 A0A2A3E4L6 B3MV29 E2AAB5 B4P2B2 I4DL23 B0W8T2 A0A1W4UQE9 A0A0P8ZHW2 A0A0R1DN25 A0A158NQG5 A0A1Q3F214 A0A1Q3F1I1 B3N7M4 A0A067RDJ2 B3MV32 B4JBF6 A0A1Y1M200 E0V9M0 A0A3B0JI06 A0A2J7QRU7 A0A0A1XKZ4 A0A0L0CNF3 A0A151HYM1 A0A1I8QC70 A0A0N0BJ07 B4ICU0 A0A0L7QS53 Q9VPM1 B4Q610 A0A034VG28 A0A195CAX6 Q17IV4 B4LU58 A0A3B0KE89 A0A1B6H1N2 A0A1B6DB79 W8BVL0 A0A0M4EEH4 B4KHG1 T1HT59 A0A088A3N5 A0A154NYT5 A0A1B6I9I4 A0A151WRL4 A0A1B6H0S0 B4GJP2 B5DHQ5 A0A1B6GZY2 A0A023F305 A0A195DUZ6 E9JB07 Q7Q4M3 A0A1W4WRQ0 A0A195EXM9 A0A1A9XYK2 A0A1B0BJJ5 A0A0K8VH45 A0A182HAK4 A0A182NUU9 A0A1L8DRK2 A0A1I8M936 A0A2M4CGT1 W5J422 A0A2M4CGT8 A0A182WHH0 A0A1A9UDD5 A0A182Y5J0 A0A1A9VLN0 A0A034VL44 A0A182RRK5 A0A182P3A4 N6TF58 U4U9J7 A0A182IBX4 A0A182L077 A0A1J1I0F0 A0A182LUH3 A0A182X151 A0A182VE55 A0A087UX04 A0A1A9ZZF3 A0A1B0FMX1 A0A1A9WW05 T1KAI3 A0A2P8ZNA2 A0A0N7ZBR3
A0A194QL60 A0A194PKA3 A0A2A4JNY5 A0A0L7KQ81 D6WDX9 A0A026WXE5 B4N791 A0A0A9Y6K3 A0A2A3E4L6 B3MV29 E2AAB5 B4P2B2 I4DL23 B0W8T2 A0A1W4UQE9 A0A0P8ZHW2 A0A0R1DN25 A0A158NQG5 A0A1Q3F214 A0A1Q3F1I1 B3N7M4 A0A067RDJ2 B3MV32 B4JBF6 A0A1Y1M200 E0V9M0 A0A3B0JI06 A0A2J7QRU7 A0A0A1XKZ4 A0A0L0CNF3 A0A151HYM1 A0A1I8QC70 A0A0N0BJ07 B4ICU0 A0A0L7QS53 Q9VPM1 B4Q610 A0A034VG28 A0A195CAX6 Q17IV4 B4LU58 A0A3B0KE89 A0A1B6H1N2 A0A1B6DB79 W8BVL0 A0A0M4EEH4 B4KHG1 T1HT59 A0A088A3N5 A0A154NYT5 A0A1B6I9I4 A0A151WRL4 A0A1B6H0S0 B4GJP2 B5DHQ5 A0A1B6GZY2 A0A023F305 A0A195DUZ6 E9JB07 Q7Q4M3 A0A1W4WRQ0 A0A195EXM9 A0A1A9XYK2 A0A1B0BJJ5 A0A0K8VH45 A0A182HAK4 A0A182NUU9 A0A1L8DRK2 A0A1I8M936 A0A2M4CGT1 W5J422 A0A2M4CGT8 A0A182WHH0 A0A1A9UDD5 A0A182Y5J0 A0A1A9VLN0 A0A034VL44 A0A182RRK5 A0A182P3A4 N6TF58 U4U9J7 A0A182IBX4 A0A182L077 A0A1J1I0F0 A0A182LUH3 A0A182X151 A0A182VE55 A0A087UX04 A0A1A9ZZF3 A0A1B0FMX1 A0A1A9WW05 T1KAI3 A0A2P8ZNA2 A0A0N7ZBR3
EC Number
2.3.1.225
Pubmed
19121390
23622113
22118469
26354079
26227816
18362917
+ More
19820115 24508170 17994087 25401762 20798317 17550304 22651552 21347285 18057021 24845553 28004739 20566863 25830018 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 17510324 24495485 15632085 23185243 25474469 21282665 12364791 26483478 25315136 20920257 23761445 25244985 23537049 20966253 29403074
19820115 24508170 17994087 25401762 20798317 17550304 22651552 21347285 18057021 24845553 28004739 20566863 25830018 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 17510324 24495485 15632085 23185243 25474469 21282665 12364791 26483478 25315136 20920257 23761445 25244985 23537049 20966253 29403074
EMBL
BABH01042040
ODYU01001529
SOQ37870.1
NWSH01001013
PCG73131.1
RSAL01000016
+ More
RVE53065.1 GAIX01012051 JAA80509.1 AGBW02012957 OWR44162.1 KQ461198 KPJ06119.1 KQ459606 KPI91525.1 PCG73130.1 JTDY01007413 KOB65216.1 KQ971324 EFA01195.1 KK107078 EZA60503.1 CH964182 EDW80232.1 GBHO01018475 GBHO01018474 GBRD01005718 JAG25129.1 JAG25130.1 JAG60103.1 KZ288403 PBC26206.1 CH902624 EDV33094.1 GL438061 EFN69626.1 CM000157 EDW87108.1 KRJ97027.1 AK401991 BAM18613.1 DS231860 EDS39311.1 KPU74294.1 KRJ97026.1 ADTU01023314 GFDL01013449 JAV21596.1 GFDL01013633 JAV21412.1 CH954177 EDV57200.1 KQS69917.1 KK852529 KDR21946.1 EDV33097.1 CH916368 EDW03979.1 GEZM01042426 JAV79864.1 AAZO01000194 DS234994 EEB10076.1 OUUW01000006 SPP81977.1 NEVH01011885 PNF31297.1 GBXI01002631 JAD11661.1 JRES01000254 KNC32954.1 KQ976723 KYM76686.1 KQ435724 KOX78504.1 CH480829 EDW45366.1 KQ414758 KOC61462.1 AE014134 AY051946 BT124833 AAF51525.2 AAK93370.1 AAN10512.1 ADF87928.1 CM000361 CM002910 EDX03194.1 KMY87272.1 GAKP01016716 JAC42236.1 KQ978023 KYM98014.1 CH477236 EAT46639.1 CH940649 EDW64045.1 SPP81978.1 GECZ01001188 JAS68581.1 GEDC01014349 JAS22949.1 GAMC01003318 JAC03238.1 CP012523 ALC38542.1 CH933807 EDW11225.1 KRG02584.1 ACPB03006071 KQ434783 KZC04732.1 GECU01024134 JAS83572.1 KQ982803 KYQ50549.1 GECZ01001495 JAS68274.1 CH479184 EDW36858.1 CH379058 EDY69832.1 KRT03619.1 GECZ01001802 JAS67967.1 GBBI01002977 JAC15735.1 KQ980322 KYN16572.1 GL770707 EFZ09996.1 AAAB01008964 EAA12777.4 KQ981948 KYN32652.1 JXJN01015425 JXJN01015426 GDHF01014449 JAI37865.1 JXUM01122850 KQ566650 KXJ69942.1 GFDF01005077 JAV09007.1 GGFL01000345 MBW64523.1 ADMH02002168 ETN58083.1 GGFL01000346 MBW64524.1 GAKP01016714 JAC42238.1 APGK01030352 KB740770 ENN78999.1 KB632132 ERL89003.1 APCN01000754 CVRI01000025 CRK92302.1 AXCM01010375 AXCM01010376 KK122085 KFM81893.1 CCAG010004915 CAEY01001939 PYGN01000012 PSN57963.1 GDRN01079617 JAI62368.1
RVE53065.1 GAIX01012051 JAA80509.1 AGBW02012957 OWR44162.1 KQ461198 KPJ06119.1 KQ459606 KPI91525.1 PCG73130.1 JTDY01007413 KOB65216.1 KQ971324 EFA01195.1 KK107078 EZA60503.1 CH964182 EDW80232.1 GBHO01018475 GBHO01018474 GBRD01005718 JAG25129.1 JAG25130.1 JAG60103.1 KZ288403 PBC26206.1 CH902624 EDV33094.1 GL438061 EFN69626.1 CM000157 EDW87108.1 KRJ97027.1 AK401991 BAM18613.1 DS231860 EDS39311.1 KPU74294.1 KRJ97026.1 ADTU01023314 GFDL01013449 JAV21596.1 GFDL01013633 JAV21412.1 CH954177 EDV57200.1 KQS69917.1 KK852529 KDR21946.1 EDV33097.1 CH916368 EDW03979.1 GEZM01042426 JAV79864.1 AAZO01000194 DS234994 EEB10076.1 OUUW01000006 SPP81977.1 NEVH01011885 PNF31297.1 GBXI01002631 JAD11661.1 JRES01000254 KNC32954.1 KQ976723 KYM76686.1 KQ435724 KOX78504.1 CH480829 EDW45366.1 KQ414758 KOC61462.1 AE014134 AY051946 BT124833 AAF51525.2 AAK93370.1 AAN10512.1 ADF87928.1 CM000361 CM002910 EDX03194.1 KMY87272.1 GAKP01016716 JAC42236.1 KQ978023 KYM98014.1 CH477236 EAT46639.1 CH940649 EDW64045.1 SPP81978.1 GECZ01001188 JAS68581.1 GEDC01014349 JAS22949.1 GAMC01003318 JAC03238.1 CP012523 ALC38542.1 CH933807 EDW11225.1 KRG02584.1 ACPB03006071 KQ434783 KZC04732.1 GECU01024134 JAS83572.1 KQ982803 KYQ50549.1 GECZ01001495 JAS68274.1 CH479184 EDW36858.1 CH379058 EDY69832.1 KRT03619.1 GECZ01001802 JAS67967.1 GBBI01002977 JAC15735.1 KQ980322 KYN16572.1 GL770707 EFZ09996.1 AAAB01008964 EAA12777.4 KQ981948 KYN32652.1 JXJN01015425 JXJN01015426 GDHF01014449 JAI37865.1 JXUM01122850 KQ566650 KXJ69942.1 GFDF01005077 JAV09007.1 GGFL01000345 MBW64523.1 ADMH02002168 ETN58083.1 GGFL01000346 MBW64524.1 GAKP01016714 JAC42238.1 APGK01030352 KB740770 ENN78999.1 KB632132 ERL89003.1 APCN01000754 CVRI01000025 CRK92302.1 AXCM01010375 AXCM01010376 KK122085 KFM81893.1 CCAG010004915 CAEY01001939 PYGN01000012 PSN57963.1 GDRN01079617 JAI62368.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000037510 UP000007266 UP000053097 UP000007798 UP000242457 UP000007801 UP000000311 UP000002282 UP000002320 UP000192221 UP000005205 UP000008711 UP000027135 UP000001070 UP000009046 UP000268350 UP000235965 UP000037069 UP000078540 UP000095300 UP000053105 UP000001292 UP000053825 UP000000803 UP000000304 UP000078542 UP000008820 UP000008792 UP000092553 UP000009192 UP000015103 UP000005203 UP000076502 UP000075809 UP000008744 UP000001819 UP000078492 UP000007062 UP000192223 UP000078541 UP000092443 UP000092460 UP000069940 UP000249989 UP000075884 UP000095301 UP000000673 UP000075920 UP000078200 UP000076408 UP000075900 UP000075885 UP000019118 UP000030742 UP000075840 UP000075882 UP000183832 UP000075883 UP000076407 UP000075903 UP000054359 UP000092445 UP000092444 UP000091820 UP000015104 UP000245037
UP000037510 UP000007266 UP000053097 UP000007798 UP000242457 UP000007801 UP000000311 UP000002282 UP000002320 UP000192221 UP000005205 UP000008711 UP000027135 UP000001070 UP000009046 UP000268350 UP000235965 UP000037069 UP000078540 UP000095300 UP000053105 UP000001292 UP000053825 UP000000803 UP000000304 UP000078542 UP000008820 UP000008792 UP000092553 UP000009192 UP000015103 UP000005203 UP000076502 UP000075809 UP000008744 UP000001819 UP000078492 UP000007062 UP000192223 UP000078541 UP000092443 UP000092460 UP000069940 UP000249989 UP000075884 UP000095301 UP000000673 UP000075920 UP000078200 UP000076408 UP000075900 UP000075885 UP000019118 UP000030742 UP000075840 UP000075882 UP000183832 UP000075883 UP000076407 UP000075903 UP000054359 UP000092445 UP000092444 UP000091820 UP000015104 UP000245037
Interpro
Gene 3D
ProteinModelPortal
H9JBA2
A0A2H1VAJ1
A0A2A4JMB6
A0A3S2LSS1
S4NTX3
A0A212ERP2
+ More
A0A194QL60 A0A194PKA3 A0A2A4JNY5 A0A0L7KQ81 D6WDX9 A0A026WXE5 B4N791 A0A0A9Y6K3 A0A2A3E4L6 B3MV29 E2AAB5 B4P2B2 I4DL23 B0W8T2 A0A1W4UQE9 A0A0P8ZHW2 A0A0R1DN25 A0A158NQG5 A0A1Q3F214 A0A1Q3F1I1 B3N7M4 A0A067RDJ2 B3MV32 B4JBF6 A0A1Y1M200 E0V9M0 A0A3B0JI06 A0A2J7QRU7 A0A0A1XKZ4 A0A0L0CNF3 A0A151HYM1 A0A1I8QC70 A0A0N0BJ07 B4ICU0 A0A0L7QS53 Q9VPM1 B4Q610 A0A034VG28 A0A195CAX6 Q17IV4 B4LU58 A0A3B0KE89 A0A1B6H1N2 A0A1B6DB79 W8BVL0 A0A0M4EEH4 B4KHG1 T1HT59 A0A088A3N5 A0A154NYT5 A0A1B6I9I4 A0A151WRL4 A0A1B6H0S0 B4GJP2 B5DHQ5 A0A1B6GZY2 A0A023F305 A0A195DUZ6 E9JB07 Q7Q4M3 A0A1W4WRQ0 A0A195EXM9 A0A1A9XYK2 A0A1B0BJJ5 A0A0K8VH45 A0A182HAK4 A0A182NUU9 A0A1L8DRK2 A0A1I8M936 A0A2M4CGT1 W5J422 A0A2M4CGT8 A0A182WHH0 A0A1A9UDD5 A0A182Y5J0 A0A1A9VLN0 A0A034VL44 A0A182RRK5 A0A182P3A4 N6TF58 U4U9J7 A0A182IBX4 A0A182L077 A0A1J1I0F0 A0A182LUH3 A0A182X151 A0A182VE55 A0A087UX04 A0A1A9ZZF3 A0A1B0FMX1 A0A1A9WW05 T1KAI3 A0A2P8ZNA2 A0A0N7ZBR3
A0A194QL60 A0A194PKA3 A0A2A4JNY5 A0A0L7KQ81 D6WDX9 A0A026WXE5 B4N791 A0A0A9Y6K3 A0A2A3E4L6 B3MV29 E2AAB5 B4P2B2 I4DL23 B0W8T2 A0A1W4UQE9 A0A0P8ZHW2 A0A0R1DN25 A0A158NQG5 A0A1Q3F214 A0A1Q3F1I1 B3N7M4 A0A067RDJ2 B3MV32 B4JBF6 A0A1Y1M200 E0V9M0 A0A3B0JI06 A0A2J7QRU7 A0A0A1XKZ4 A0A0L0CNF3 A0A151HYM1 A0A1I8QC70 A0A0N0BJ07 B4ICU0 A0A0L7QS53 Q9VPM1 B4Q610 A0A034VG28 A0A195CAX6 Q17IV4 B4LU58 A0A3B0KE89 A0A1B6H1N2 A0A1B6DB79 W8BVL0 A0A0M4EEH4 B4KHG1 T1HT59 A0A088A3N5 A0A154NYT5 A0A1B6I9I4 A0A151WRL4 A0A1B6H0S0 B4GJP2 B5DHQ5 A0A1B6GZY2 A0A023F305 A0A195DUZ6 E9JB07 Q7Q4M3 A0A1W4WRQ0 A0A195EXM9 A0A1A9XYK2 A0A1B0BJJ5 A0A0K8VH45 A0A182HAK4 A0A182NUU9 A0A1L8DRK2 A0A1I8M936 A0A2M4CGT1 W5J422 A0A2M4CGT8 A0A182WHH0 A0A1A9UDD5 A0A182Y5J0 A0A1A9VLN0 A0A034VL44 A0A182RRK5 A0A182P3A4 N6TF58 U4U9J7 A0A182IBX4 A0A182L077 A0A1J1I0F0 A0A182LUH3 A0A182X151 A0A182VE55 A0A087UX04 A0A1A9ZZF3 A0A1B0FMX1 A0A1A9WW05 T1KAI3 A0A2P8ZNA2 A0A0N7ZBR3
PDB
4XP7
E-value=1.35675e-26,
Score=298
Ontologies
GO
Topology
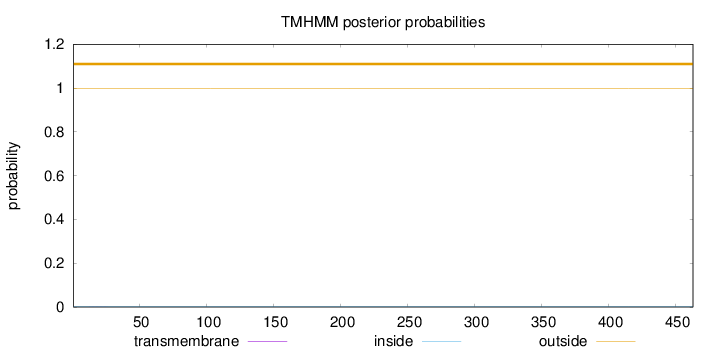
Length:
463
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00273
Exp number, first 60 AAs:
0.00273
Total prob of N-in:
0.00279
outside
1 - 463
Population Genetic Test Statistics
Pi
217.738305
Theta
162.624417
Tajima's D
1.079118
CLR
0
CSRT
0.68341582920854
Interpretation
Uncertain
