Gene
KWMTBOMO05653
Pre Gene Modal
BGIBMGA006790
Annotation
PREDICTED:_A-kinase_anchor_protein_10?_mitochondrial_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.228
Sequence
CDS
ATGATAAAGTTTTGGAAAAGTGCAGCAAAGGGCAAAAGCGGGTGCCCTGTGCGACCTCCTGAGAAGACTTCTGAAACTACTGTTGATTTCAATAACATACTAAGTAGTACGCAATTTCTTCATGACATTAAAGATTCTGAAGATATTTTTGAAAAGAAATCCAAGTTGTCGTTGTCATTAAATGACACTCTCCTTAAAAGAAAAGCAACTACGTATTTTATGCAATACATGGAAACCATCGGACAACATTTACTTGTTAAATGTTGGCTAGATTTGGAAGATTTCAAATCACACTTTAGAAATACAGACAATGTAGTGCTAAATATGTACGGGAATAATAAAAAATCTCAAAATTCCACTGAAGAAGCTGTAAATTTATTCAAAAAGTATATAGCTTTAGAAGCACCATATCAGATAGATATACCTGATAATGTAAGAAAATCAGTGATAACAAATATTTGTCATGTGGATGGACCGACTGAAGACTGTTTTAAGTCAGTGCAAGATATGCTATTTACTCGAATCGAAGAAAATCATTTTGGTGACTTCCTCAATACTCCACTTTATATCAAAGCACAAATAGATATACTCACAAATGGCTCTCTCCAATTGAAAGATATTTTATACAATGACACACTCTTATTTTACTTCACTGAATATTTGGAACAAGAATCTTGTAGGATATTGCTTGAATTTTTAATGGCCATTATACATTTCAGAGATAACTTAATCAATAATAATAATGATAATGCTGATCAAGCTCAAGCTGACGCCATTGTAATATACGAAAAATACTTTTCCTTACAAGCAACTAACCCACTTGGTTTCCCAACAGATATTAGACTTAAAATCGAGAGTGAAATATGTAGAGAAAACGGACCTCAGTATGATTGTTTTGATCTGCCTTATCGAATAGTTTATATGACAGTTACCCATTATGTGAAAACATTTCTTGAATCCAATATGTACTATAAATATCTTTCAGAAATGATCCACTCTGTGGATTATAATATGTCATCGCTAAATCATAAGTCTTATTCTGACTGTAGCAGTGAATATAGTGTCAGTACACAAAATACATTGCTGGCAATGGGGGATCCAGTCTTCCGAAGAAAAAAACGCAACCACTCTGTACCTGATATGACGATAGACTCTAATCAGCTTTACAATGCAGATTCACTATGGAAAAGAACTAGACATGAAGGCCTTACTTTAGGAAGAATAAATAGTTTAGGTAGGTTTGAATCCAAGTTTGAACCAGATCCAGACAAAAAAGAATCTGTTTTAAAAAAAATGGTAAATCGCTTTGTGCCAATAAGTACATCTAAAGTAGAAGAAGAAATGGCTTGGCAAATAGCCCACATGATTGTAAAAGATGTAACAGATCTGACTATGGCACCACCAGATAATGAAGAAGAAGAAATGTAA
Protein
MIKFWKSAAKGKSGCPVRPPEKTSETTVDFNNILSSTQFLHDIKDSEDIFEKKSKLSLSLNDTLLKRKATTYFMQYMETIGQHLLVKCWLDLEDFKSHFRNTDNVVLNMYGNNKKSQNSTEEAVNLFKKYIALEAPYQIDIPDNVRKSVITNICHVDGPTEDCFKSVQDMLFTRIEENHFGDFLNTPLYIKAQIDILTNGSLQLKDILYNDTLLFYFTEYLEQESCRILLEFLMAIIHFRDNLINNNNDNADQAQADAIVIYEKYFSLQATNPLGFPTDIRLKIESEICRENGPQYDCFDLPYRIVYMTVTHYVKTFLESNMYYKYLSEMIHSVDYNMSSLNHKSYSDCSSEYSVSTQNTLLAMGDPVFRRKKRNHSVPDMTIDSNQLYNADSLWKRTRHEGLTLGRINSLGRFESKFEPDPDKKESVLKKMVNRFVPISTSKVEEEMAWQIAHMIVKDVTDLTMAPPDNEEEEM
Summary
Uniprot
H9JB95
A0A0L7L3L3
A0A2A4J712
A0A2H1W549
A0A1E1WSW0
A0A194PKB4
+ More
A0A212ERU8 A0A1W4XWN7 D6X0R7 A0A2J7Q8L5 N6SZT1 A0A1Y1K6R9 A0A158NCN9 E0VIT2 A0A154P399 A0A195BWS7 A0A2A3ES48 F4WFB8 E2BQ42 A0A1Z5LCM7 A0A1B0D3I0 A0A195CG26 V3ZGG6 A0A0C9R3N3 A0A0L7R1N0 A0A195ELU9 A0A151XF38 A0A088A0D9 A0A1Z5KVV9 A0A0M8ZSP5 E2A227 A0A1E1XLR0 A0A131YW31 A0A224YGF7 A0A195FA51 A0A3L8D4T4 A0A026W035 A0A1E1XDJ7 A0A293M2W4 A0A1S3JJ81 A0A3Q3AI81 A0A232F254 Q170P1 A0A2R2MIJ6 A0A3B4CJ05 A0A3B5BDZ4 A0A0L8HPT7 T1JML4 A0A3B3BR59 A0A1A7WG62 A0A3P9AL77 A0A3B4H498 A0A3B3D949 A0A3Q1HN17 A0A3Q3JL14 A0A3P8SNT3 A0A2I4BVY5 A0A3Q1CDG0 A0A3S2P3P5 A0A3P8Z5D7 A0A3B4UX73 A0A3B4Y472 C3YDJ0 A0A1A7X8G6 A0A3B4UW07 A0A1S3LQW4 A0A1A8EA04 A0A1A8BLM1 A0A1A8LJ21 A0A1A8ACB0 A0A1A8QFT8 F6WGK9 A0A3Q2DAQ3 A0A3P9ISC7 A0A1A8JGZ9 A0A1S3F697 A0A1A8FSJ9 E9QB82 A0A3B1J640 A0A3Q3LCZ2 A0A336M946 A0A3Q1J756 A0A1S3LQZ9 F1QDF6 A0A2U4C2X7 A0A3P9LCX5 A0A2D0QEQ1 A0A060WX32 A0A2D0QDP7 A0A1A7ZNA2 A0A146MWB8 A0A151NBB5 A0A1A8DSS4 A0A1A8CES9 A0A3Q3LRK9 A0A1A8JSE5 A0A210Q0E0 A0A1A8MVY1
A0A212ERU8 A0A1W4XWN7 D6X0R7 A0A2J7Q8L5 N6SZT1 A0A1Y1K6R9 A0A158NCN9 E0VIT2 A0A154P399 A0A195BWS7 A0A2A3ES48 F4WFB8 E2BQ42 A0A1Z5LCM7 A0A1B0D3I0 A0A195CG26 V3ZGG6 A0A0C9R3N3 A0A0L7R1N0 A0A195ELU9 A0A151XF38 A0A088A0D9 A0A1Z5KVV9 A0A0M8ZSP5 E2A227 A0A1E1XLR0 A0A131YW31 A0A224YGF7 A0A195FA51 A0A3L8D4T4 A0A026W035 A0A1E1XDJ7 A0A293M2W4 A0A1S3JJ81 A0A3Q3AI81 A0A232F254 Q170P1 A0A2R2MIJ6 A0A3B4CJ05 A0A3B5BDZ4 A0A0L8HPT7 T1JML4 A0A3B3BR59 A0A1A7WG62 A0A3P9AL77 A0A3B4H498 A0A3B3D949 A0A3Q1HN17 A0A3Q3JL14 A0A3P8SNT3 A0A2I4BVY5 A0A3Q1CDG0 A0A3S2P3P5 A0A3P8Z5D7 A0A3B4UX73 A0A3B4Y472 C3YDJ0 A0A1A7X8G6 A0A3B4UW07 A0A1S3LQW4 A0A1A8EA04 A0A1A8BLM1 A0A1A8LJ21 A0A1A8ACB0 A0A1A8QFT8 F6WGK9 A0A3Q2DAQ3 A0A3P9ISC7 A0A1A8JGZ9 A0A1S3F697 A0A1A8FSJ9 E9QB82 A0A3B1J640 A0A3Q3LCZ2 A0A336M946 A0A3Q1J756 A0A1S3LQZ9 F1QDF6 A0A2U4C2X7 A0A3P9LCX5 A0A2D0QEQ1 A0A060WX32 A0A2D0QDP7 A0A1A7ZNA2 A0A146MWB8 A0A151NBB5 A0A1A8DSS4 A0A1A8CES9 A0A3Q3LRK9 A0A1A8JSE5 A0A210Q0E0 A0A1A8MVY1
Pubmed
EMBL
BABH01008832
JTDY01003251
KOB69879.1
NWSH01002908
PCG67314.1
ODYU01006404
+ More
SOQ48215.1 GDQN01001037 JAT90017.1 KQ459606 KPI91535.1 AGBW02012957 OWR44174.1 KQ971372 EFA10548.1 NEVH01016952 PNF24924.1 APGK01050248 KB741169 KB632294 ENN73299.1 ERL91658.1 GEZM01093750 JAV56148.1 ADTU01012004 ADTU01012005 DS235206 EEB13288.1 KQ434809 KZC06303.1 KQ976396 KYM93037.1 KZ288194 PBC33949.1 GL888115 EGI67169.1 GL449694 EFN82230.1 GFJQ02001804 JAW05166.1 AJVK01010976 AJVK01010977 AJVK01010978 KQ977791 KYM99715.1 KB203738 ESO83252.1 GBYB01010934 JAG80701.1 KQ414667 KOC64748.1 KQ978691 KYN29255.1 KQ982215 KYQ58941.1 GFJQ02008009 JAV98960.1 KQ435863 KOX70345.1 GL435917 EFN72511.1 GFAA01003271 JAU00164.1 GEDV01005154 JAP83403.1 GFPF01005632 MAA16778.1 KQ981727 KYN36924.1 QOIP01000014 RLU15023.1 KK107609 EZA48489.1 GFAC01002012 JAT97176.1 GFWV01010584 MAA35313.1 NNAY01001197 OXU24781.1 CH477471 EAT40409.1 KQ417691 KOF90750.1 JH431878 HADW01003567 SBP04967.1 CM012450 RVE63514.1 GG666503 EEN61801.1 HADW01012966 HADX01014307 SBP14366.1 HAEA01014810 SBQ43290.1 HADZ01003585 SBP67526.1 HAEF01007136 SBR44518.1 HADY01013370 HAEJ01018947 SBP51855.1 HAEH01011352 SBR92009.1 HAED01022445 SBR09200.1 HAEB01015455 SBQ61982.1 BX511157 UFQT01000560 SSX25313.1 FR904775 CDQ71567.1 HADY01005311 HAEJ01005262 SBP43796.1 GCES01162503 JAQ23819.1 AKHW03003672 KYO33775.1 HAEA01007389 SBQ35869.1 HADZ01013622 SBP77563.1 HAED01006896 HAEE01003003 SBR23023.1 NEDP02005302 OWF42224.1 HAEF01019495 HAEG01000333 SBR60654.1
SOQ48215.1 GDQN01001037 JAT90017.1 KQ459606 KPI91535.1 AGBW02012957 OWR44174.1 KQ971372 EFA10548.1 NEVH01016952 PNF24924.1 APGK01050248 KB741169 KB632294 ENN73299.1 ERL91658.1 GEZM01093750 JAV56148.1 ADTU01012004 ADTU01012005 DS235206 EEB13288.1 KQ434809 KZC06303.1 KQ976396 KYM93037.1 KZ288194 PBC33949.1 GL888115 EGI67169.1 GL449694 EFN82230.1 GFJQ02001804 JAW05166.1 AJVK01010976 AJVK01010977 AJVK01010978 KQ977791 KYM99715.1 KB203738 ESO83252.1 GBYB01010934 JAG80701.1 KQ414667 KOC64748.1 KQ978691 KYN29255.1 KQ982215 KYQ58941.1 GFJQ02008009 JAV98960.1 KQ435863 KOX70345.1 GL435917 EFN72511.1 GFAA01003271 JAU00164.1 GEDV01005154 JAP83403.1 GFPF01005632 MAA16778.1 KQ981727 KYN36924.1 QOIP01000014 RLU15023.1 KK107609 EZA48489.1 GFAC01002012 JAT97176.1 GFWV01010584 MAA35313.1 NNAY01001197 OXU24781.1 CH477471 EAT40409.1 KQ417691 KOF90750.1 JH431878 HADW01003567 SBP04967.1 CM012450 RVE63514.1 GG666503 EEN61801.1 HADW01012966 HADX01014307 SBP14366.1 HAEA01014810 SBQ43290.1 HADZ01003585 SBP67526.1 HAEF01007136 SBR44518.1 HADY01013370 HAEJ01018947 SBP51855.1 HAEH01011352 SBR92009.1 HAED01022445 SBR09200.1 HAEB01015455 SBQ61982.1 BX511157 UFQT01000560 SSX25313.1 FR904775 CDQ71567.1 HADY01005311 HAEJ01005262 SBP43796.1 GCES01162503 JAQ23819.1 AKHW03003672 KYO33775.1 HAEA01007389 SBQ35869.1 HADZ01013622 SBP77563.1 HAED01006896 HAEE01003003 SBR23023.1 NEDP02005302 OWF42224.1 HAEF01019495 HAEG01000333 SBR60654.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000007151
UP000192223
+ More
UP000007266 UP000235965 UP000019118 UP000030742 UP000005205 UP000009046 UP000076502 UP000078540 UP000242457 UP000007755 UP000008237 UP000092462 UP000078542 UP000030746 UP000053825 UP000078492 UP000075809 UP000005203 UP000053105 UP000000311 UP000078541 UP000279307 UP000053097 UP000085678 UP000264800 UP000215335 UP000008820 UP000261440 UP000261400 UP000053454 UP000261560 UP000265140 UP000261460 UP000257200 UP000261600 UP000265080 UP000192220 UP000257160 UP000261420 UP000261360 UP000001554 UP000087266 UP000002281 UP000265020 UP000265200 UP000081671 UP000000437 UP000018467 UP000261640 UP000265040 UP000245320 UP000265180 UP000221080 UP000193380 UP000050525 UP000242188
UP000007266 UP000235965 UP000019118 UP000030742 UP000005205 UP000009046 UP000076502 UP000078540 UP000242457 UP000007755 UP000008237 UP000092462 UP000078542 UP000030746 UP000053825 UP000078492 UP000075809 UP000005203 UP000053105 UP000000311 UP000078541 UP000279307 UP000053097 UP000085678 UP000264800 UP000215335 UP000008820 UP000261440 UP000261400 UP000053454 UP000261560 UP000265140 UP000261460 UP000257200 UP000261600 UP000265080 UP000192220 UP000257160 UP000261420 UP000261360 UP000001554 UP000087266 UP000002281 UP000265020 UP000265200 UP000081671 UP000000437 UP000018467 UP000261640 UP000265040 UP000245320 UP000265180 UP000221080 UP000193380 UP000050525 UP000242188
Pfam
Interpro
IPR016137
RGS
+ More
IPR036305 RGS_sf
IPR037719 AKAP10_AKB_dom
IPR041466 Dynein_AAA5_ext
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR042219 AAA_lid_11_sf
IPR026983 DHC_fam
IPR013594 Dynein_heavy_dom-1
IPR035699 AAA_6
IPR041658 AAA_lid_11
IPR041228 Dynein_C
IPR042228 Dynein_2_C
IPR042222 Dynein_2_N
IPR035706 AAA_9
IPR041589 DNAH3_AAA_lid_1
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR036305 RGS_sf
IPR037719 AKAP10_AKB_dom
IPR041466 Dynein_AAA5_ext
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR042219 AAA_lid_11_sf
IPR026983 DHC_fam
IPR013594 Dynein_heavy_dom-1
IPR035699 AAA_6
IPR041658 AAA_lid_11
IPR041228 Dynein_C
IPR042228 Dynein_2_C
IPR042222 Dynein_2_N
IPR035706 AAA_9
IPR041589 DNAH3_AAA_lid_1
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
Gene 3D
ProteinModelPortal
H9JB95
A0A0L7L3L3
A0A2A4J712
A0A2H1W549
A0A1E1WSW0
A0A194PKB4
+ More
A0A212ERU8 A0A1W4XWN7 D6X0R7 A0A2J7Q8L5 N6SZT1 A0A1Y1K6R9 A0A158NCN9 E0VIT2 A0A154P399 A0A195BWS7 A0A2A3ES48 F4WFB8 E2BQ42 A0A1Z5LCM7 A0A1B0D3I0 A0A195CG26 V3ZGG6 A0A0C9R3N3 A0A0L7R1N0 A0A195ELU9 A0A151XF38 A0A088A0D9 A0A1Z5KVV9 A0A0M8ZSP5 E2A227 A0A1E1XLR0 A0A131YW31 A0A224YGF7 A0A195FA51 A0A3L8D4T4 A0A026W035 A0A1E1XDJ7 A0A293M2W4 A0A1S3JJ81 A0A3Q3AI81 A0A232F254 Q170P1 A0A2R2MIJ6 A0A3B4CJ05 A0A3B5BDZ4 A0A0L8HPT7 T1JML4 A0A3B3BR59 A0A1A7WG62 A0A3P9AL77 A0A3B4H498 A0A3B3D949 A0A3Q1HN17 A0A3Q3JL14 A0A3P8SNT3 A0A2I4BVY5 A0A3Q1CDG0 A0A3S2P3P5 A0A3P8Z5D7 A0A3B4UX73 A0A3B4Y472 C3YDJ0 A0A1A7X8G6 A0A3B4UW07 A0A1S3LQW4 A0A1A8EA04 A0A1A8BLM1 A0A1A8LJ21 A0A1A8ACB0 A0A1A8QFT8 F6WGK9 A0A3Q2DAQ3 A0A3P9ISC7 A0A1A8JGZ9 A0A1S3F697 A0A1A8FSJ9 E9QB82 A0A3B1J640 A0A3Q3LCZ2 A0A336M946 A0A3Q1J756 A0A1S3LQZ9 F1QDF6 A0A2U4C2X7 A0A3P9LCX5 A0A2D0QEQ1 A0A060WX32 A0A2D0QDP7 A0A1A7ZNA2 A0A146MWB8 A0A151NBB5 A0A1A8DSS4 A0A1A8CES9 A0A3Q3LRK9 A0A1A8JSE5 A0A210Q0E0 A0A1A8MVY1
A0A212ERU8 A0A1W4XWN7 D6X0R7 A0A2J7Q8L5 N6SZT1 A0A1Y1K6R9 A0A158NCN9 E0VIT2 A0A154P399 A0A195BWS7 A0A2A3ES48 F4WFB8 E2BQ42 A0A1Z5LCM7 A0A1B0D3I0 A0A195CG26 V3ZGG6 A0A0C9R3N3 A0A0L7R1N0 A0A195ELU9 A0A151XF38 A0A088A0D9 A0A1Z5KVV9 A0A0M8ZSP5 E2A227 A0A1E1XLR0 A0A131YW31 A0A224YGF7 A0A195FA51 A0A3L8D4T4 A0A026W035 A0A1E1XDJ7 A0A293M2W4 A0A1S3JJ81 A0A3Q3AI81 A0A232F254 Q170P1 A0A2R2MIJ6 A0A3B4CJ05 A0A3B5BDZ4 A0A0L8HPT7 T1JML4 A0A3B3BR59 A0A1A7WG62 A0A3P9AL77 A0A3B4H498 A0A3B3D949 A0A3Q1HN17 A0A3Q3JL14 A0A3P8SNT3 A0A2I4BVY5 A0A3Q1CDG0 A0A3S2P3P5 A0A3P8Z5D7 A0A3B4UX73 A0A3B4Y472 C3YDJ0 A0A1A7X8G6 A0A3B4UW07 A0A1S3LQW4 A0A1A8EA04 A0A1A8BLM1 A0A1A8LJ21 A0A1A8ACB0 A0A1A8QFT8 F6WGK9 A0A3Q2DAQ3 A0A3P9ISC7 A0A1A8JGZ9 A0A1S3F697 A0A1A8FSJ9 E9QB82 A0A3B1J640 A0A3Q3LCZ2 A0A336M946 A0A3Q1J756 A0A1S3LQZ9 F1QDF6 A0A2U4C2X7 A0A3P9LCX5 A0A2D0QEQ1 A0A060WX32 A0A2D0QDP7 A0A1A7ZNA2 A0A146MWB8 A0A151NBB5 A0A1A8DSS4 A0A1A8CES9 A0A3Q3LRK9 A0A1A8JSE5 A0A210Q0E0 A0A1A8MVY1
PDB
3C7L
E-value=0.0037808,
Score=96
Ontologies
KEGG
GO
PANTHER
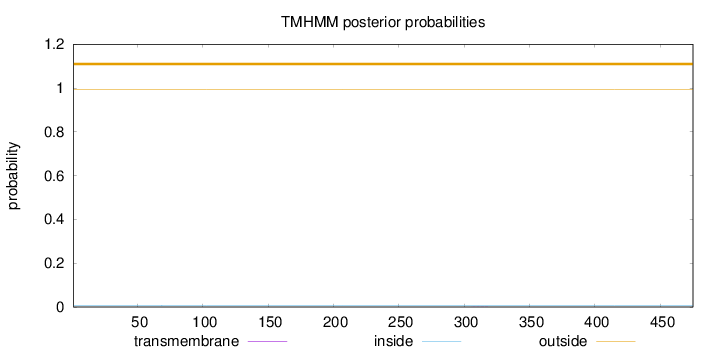
Topology
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.005
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00744
outside
1 - 475
Population Genetic Test Statistics
Pi
224.989288
Theta
168.537498
Tajima's D
0.779299
CLR
0
CSRT
0.591170441477926
Interpretation
Uncertain
