Gene
KWMTBOMO05651 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006789
Annotation
PREDICTED:_malectin-A_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.775
Sequence
CDS
ATGTCGCGACTTATCCGCATTTTTATTTCTTTACTATGTGTCATTAAAACTGCTACGGGATTAGGTGAAATCATCTACGCCATAAATGCCGGAGGTCCTGCTCATACAGACATCTACGGTATTCATTATGAAAAAGATCCATTACAGGGACGAATTGGCACTGCATCAGATTATGGTAAACAATTGGTTATGATCGGCAGAGTTAATCCTAAAGATGAAATACTTTATCAAACTGAAAGGTATCACCACAGCACGTTCGGCTACGACATTCCGGCAAACAAAGACGGCGACTATGTTTTAGTTTTAAAGTTCAGTGAAGTGTATTTCAGTTCCCCAAACATGAAAGTATTTGATGTCGTCTTGAATGGAGACCATACTATAGTAGCCGATTTGGATATTTTTGATAAAGTAGGAAGAGGTGTTGCTCATGATGAATATATACCTTACACCATAAAGAACGGGAAACTCTACTATAATGAAGAAGAATCTGATATTCGGGGAGGAAAAATTAAAGTTGAATTTATCAAAGGATATCGGGATAACCCAAAGATAAATGCTCTATATGTTATGACAGGCACAATAGATGAAGTTCCCAAATTACCACCTTTGGTTTGGCCTGAAAATGAAACTGAAGAAGCAAGAGATGAAATTGAAGAAAAGAACACCAAAGCGAGACGTGCTAGTGGGCCAAAACAACCAGACCCATATGTTATGGATGAAAGCTCAGTTTTGATTCCAGTTTTCATAACAATTGGAGCATTTATACCTTTGTTATTCTGTCTTTGTAAATTATAA
Protein
MSRLIRIFISLLCVIKTATGLGEIIYAINAGGPAHTDIYGIHYEKDPLQGRIGTASDYGKQLVMIGRVNPKDEILYQTERYHHSTFGYDIPANKDGDYVLVLKFSEVYFSSPNMKVFDVVLNGDHTIVADLDIFDKVGRGVAHDEYIPYTIKNGKLYYNEEESDIRGGKIKVEFIKGYRDNPKINALYVMTGTIDEVPKLPPLVWPENETEEARDEIEEKNTKARRASGPKQPDPYVMDESSVLIPVFITIGAFIPLLFCLCKL
Summary
Uniprot
H9JB94
A0A2A4J7B0
A0A2H1X361
A0A1E1WTB3
A0A0L7L2Z5
A0A194PE48
+ More
S4Q007 A0A2R7X5N5 A0A0A9YIW4 A0A067QKD5 A0A023FAM5 A0A224XVF6 R4G3H4 E0VMW4 A0A1B6DUX0 A0A2J7RBF7 A0A212ERP5 A0A232FAP1 D6X2Z1 A0A151WT91 E2AHG0 A0A0L7R1B2 A0A194QKS9 A0A026WDQ9 A0A2A3EHG7 A0A158P1Y9 A0A0C9RJU1 K7IQ78 A0A195FIF8 A0A1Y1KYB5 F4W665 E2BED3 A0A151IKI3 A0A195DMK1 T1IK58 A0A087UEZ7 A0A0T6B0S6 A0A087ZPN7 A0A1W4XC72 A0A0M9A408 A0A336LAH7 A0A336LE26 A0A2P8ZIZ5 A0A023FP57 A0A293N757 A0A0N7ZC60 A0A0K2TR34 A0A023FY87 G3MT06 B0W522 A0A023GP25 A0A1Q3FQN9 C1C2E4 Q17BA6 L7M877 A0A1D2N7P4 C1BRJ2 A0A023EPQ3 A0A1B0C927 A0A182H108 A0A2T7PDX7 A0A1S3D6Z9 A0A154NY41 A0A310SKQ4 E9H405 A0A0P5P9U2 A0A0P6HQS2 A0A131YPC0 A0A0P5ZMB7 A0A0P5CJL4 A0A1L8DLR5 A0A1L8DJS8 V5H0M4 A0A131YAK0 V5I1P0 V4AVB5 B7Q5I2 U5EJ84 A0A1B0D2G8 A0A0B7ADM8 C3YVX3 A0A0A1X9Z8 A0A0K8V6C0 A0A0P6BVP7 A0A2C9JGE2 A0A1S3IK20 J3JYP3 A0A385UIK0 A0A210QMS2 A0A1J1ING9 A0A3M6UBE1 G3TDG2 B7PB81 F7CD34 A0A0C9R6N9 A0A0P6CTJ9 D2I156 G1U070 A0A3Q7Y2P9 M3VYF8
S4Q007 A0A2R7X5N5 A0A0A9YIW4 A0A067QKD5 A0A023FAM5 A0A224XVF6 R4G3H4 E0VMW4 A0A1B6DUX0 A0A2J7RBF7 A0A212ERP5 A0A232FAP1 D6X2Z1 A0A151WT91 E2AHG0 A0A0L7R1B2 A0A194QKS9 A0A026WDQ9 A0A2A3EHG7 A0A158P1Y9 A0A0C9RJU1 K7IQ78 A0A195FIF8 A0A1Y1KYB5 F4W665 E2BED3 A0A151IKI3 A0A195DMK1 T1IK58 A0A087UEZ7 A0A0T6B0S6 A0A087ZPN7 A0A1W4XC72 A0A0M9A408 A0A336LAH7 A0A336LE26 A0A2P8ZIZ5 A0A023FP57 A0A293N757 A0A0N7ZC60 A0A0K2TR34 A0A023FY87 G3MT06 B0W522 A0A023GP25 A0A1Q3FQN9 C1C2E4 Q17BA6 L7M877 A0A1D2N7P4 C1BRJ2 A0A023EPQ3 A0A1B0C927 A0A182H108 A0A2T7PDX7 A0A1S3D6Z9 A0A154NY41 A0A310SKQ4 E9H405 A0A0P5P9U2 A0A0P6HQS2 A0A131YPC0 A0A0P5ZMB7 A0A0P5CJL4 A0A1L8DLR5 A0A1L8DJS8 V5H0M4 A0A131YAK0 V5I1P0 V4AVB5 B7Q5I2 U5EJ84 A0A1B0D2G8 A0A0B7ADM8 C3YVX3 A0A0A1X9Z8 A0A0K8V6C0 A0A0P6BVP7 A0A2C9JGE2 A0A1S3IK20 J3JYP3 A0A385UIK0 A0A210QMS2 A0A1J1ING9 A0A3M6UBE1 G3TDG2 B7PB81 F7CD34 A0A0C9R6N9 A0A0P6CTJ9 D2I156 G1U070 A0A3Q7Y2P9 M3VYF8
Pubmed
19121390
26227816
26354079
23622113
25401762
24845553
+ More
25474469 20566863 22118469 28648823 18362917 19820115 20798317 24508170 30249741 21347285 20075255 28004739 21719571 29403074 22216098 17510324 25576852 27289101 24945155 26483478 21292972 26830274 25765539 23254933 18563158 25830018 15562597 22516182 23537049 28812685 30382153 20431018 26131772 20010809 21993624 17975172
25474469 20566863 22118469 28648823 18362917 19820115 20798317 24508170 30249741 21347285 20075255 28004739 21719571 29403074 22216098 17510324 25576852 27289101 24945155 26483478 21292972 26830274 25765539 23254933 18563158 25830018 15562597 22516182 23537049 28812685 30382153 20431018 26131772 20010809 21993624 17975172
EMBL
BABH01008830
NWSH01002908
PCG67312.1
ODYU01013071
SOQ59745.1
GDQN01000814
+ More
JAT90240.1 JTDY01003251 KOB69878.1 KQ459606 KPI91537.1 GAIX01000618 JAA91942.1 KK857362 PTY27097.1 GBHO01010585 GBHO01010582 GBHO01010576 JAG33019.1 JAG33022.1 JAG33028.1 KK853247 KDR09386.1 GBBI01000628 JAC18084.1 GFTR01004333 JAW12093.1 ACPB03016356 GAHY01001676 JAA75834.1 DS235327 EEB14720.1 GEDC01028296 GEDC01008123 GEDC01007863 GEDC01007519 JAS09002.1 JAS29175.1 JAS29435.1 JAS29779.1 NEVH01005906 PNF38151.1 AGBW02012957 OWR44176.1 NNAY01000613 OXU27397.1 KQ971372 EFA09821.2 KQ982763 KYQ51011.1 GL439539 EFN67115.1 KQ414668 KOC64581.1 KQ461198 KPJ06132.1 KK107293 QOIP01000005 EZA53154.1 RLU22637.1 KZ288254 PBC30722.1 ADTU01006918 GBYB01004943 GBYB01013546 GBYB01013547 JAG74710.1 JAG83313.1 JAG83314.1 KQ981523 KYN40455.1 GEZM01074989 JAV64356.1 GL887707 EGI70200.1 GL447768 EFN85978.1 KQ977199 KYN04991.1 KQ980724 KYN14125.1 JH430421 KK119525 KFM75936.1 LJIG01016314 KRT80972.1 KQ435763 KOX75409.1 UFQS01002377 UFQT01002377 SSX13911.1 SSX33330.1 UFQS01002864 UFQT01002864 SSX14795.1 SSX34186.1 PYGN01000041 PSN56478.1 GBBK01001832 JAC22650.1 GFWV01022733 MAA47460.1 GDRN01072489 JAI63544.1 HACA01010914 HACA01010915 CDW28275.1 GBBL01001645 JAC25675.1 JO845007 AEO36624.1 DS231840 EDS34588.1 GBBM01000260 JAC35158.1 GFDL01005166 JAV29879.1 BT081023 ACO15447.1 CH477324 EAT43540.1 GACK01005715 JAA59319.1 LJIJ01000163 ODN01278.1 BT077221 ACO11645.1 GAPW01003094 JAC10504.1 AJWK01001788 JXUM01102368 KQ564722 KXJ71843.1 PZQS01000004 PVD31622.1 KQ434782 KZC04537.1 KQ761824 OAD56764.1 GL732589 EFX73624.1 GDIQ01141146 JAL10580.1 GDIQ01015558 JAN79179.1 GEDV01008806 JAP79751.1 GDIP01042872 LRGB01003032 JAM60843.1 KZS04876.1 GDIP01169636 JAJ53766.1 GFDF01006769 JAV07315.1 GFDF01007366 JAV06718.1 GANP01014059 JAB70409.1 GEFM01000284 JAP75512.1 GANP01001231 JAB83237.1 KB201304 ESO97776.1 ABJB010531888 ABJB010904798 ABJB010920537 ABJB010953177 DS861388 EEC14104.1 GANO01002327 JAB57544.1 AJVK01022796 HACG01031276 CEK78141.1 GG666558 EEN55537.1 GBXI01006370 JAD07922.1 GDHF01018046 JAI34268.1 GDIP01016840 JAM86875.1 APGK01043395 BT128373 KB741014 KB632204 AEE63331.1 ENN75458.1 ERL90104.1 MG546685 AYB71126.1 NEDP02002833 OWF50001.1 CVRI01000055 CRL01100.1 RCHS01001867 RMX50980.1 ABJB010645197 DS674754 EEC03853.1 AAMC01008482 GBZX01000125 JAG92615.1 GDIQ01087961 JAN06776.1 GL193954 EFB23139.1 AAGW02063829 AANG04002472
JAT90240.1 JTDY01003251 KOB69878.1 KQ459606 KPI91537.1 GAIX01000618 JAA91942.1 KK857362 PTY27097.1 GBHO01010585 GBHO01010582 GBHO01010576 JAG33019.1 JAG33022.1 JAG33028.1 KK853247 KDR09386.1 GBBI01000628 JAC18084.1 GFTR01004333 JAW12093.1 ACPB03016356 GAHY01001676 JAA75834.1 DS235327 EEB14720.1 GEDC01028296 GEDC01008123 GEDC01007863 GEDC01007519 JAS09002.1 JAS29175.1 JAS29435.1 JAS29779.1 NEVH01005906 PNF38151.1 AGBW02012957 OWR44176.1 NNAY01000613 OXU27397.1 KQ971372 EFA09821.2 KQ982763 KYQ51011.1 GL439539 EFN67115.1 KQ414668 KOC64581.1 KQ461198 KPJ06132.1 KK107293 QOIP01000005 EZA53154.1 RLU22637.1 KZ288254 PBC30722.1 ADTU01006918 GBYB01004943 GBYB01013546 GBYB01013547 JAG74710.1 JAG83313.1 JAG83314.1 KQ981523 KYN40455.1 GEZM01074989 JAV64356.1 GL887707 EGI70200.1 GL447768 EFN85978.1 KQ977199 KYN04991.1 KQ980724 KYN14125.1 JH430421 KK119525 KFM75936.1 LJIG01016314 KRT80972.1 KQ435763 KOX75409.1 UFQS01002377 UFQT01002377 SSX13911.1 SSX33330.1 UFQS01002864 UFQT01002864 SSX14795.1 SSX34186.1 PYGN01000041 PSN56478.1 GBBK01001832 JAC22650.1 GFWV01022733 MAA47460.1 GDRN01072489 JAI63544.1 HACA01010914 HACA01010915 CDW28275.1 GBBL01001645 JAC25675.1 JO845007 AEO36624.1 DS231840 EDS34588.1 GBBM01000260 JAC35158.1 GFDL01005166 JAV29879.1 BT081023 ACO15447.1 CH477324 EAT43540.1 GACK01005715 JAA59319.1 LJIJ01000163 ODN01278.1 BT077221 ACO11645.1 GAPW01003094 JAC10504.1 AJWK01001788 JXUM01102368 KQ564722 KXJ71843.1 PZQS01000004 PVD31622.1 KQ434782 KZC04537.1 KQ761824 OAD56764.1 GL732589 EFX73624.1 GDIQ01141146 JAL10580.1 GDIQ01015558 JAN79179.1 GEDV01008806 JAP79751.1 GDIP01042872 LRGB01003032 JAM60843.1 KZS04876.1 GDIP01169636 JAJ53766.1 GFDF01006769 JAV07315.1 GFDF01007366 JAV06718.1 GANP01014059 JAB70409.1 GEFM01000284 JAP75512.1 GANP01001231 JAB83237.1 KB201304 ESO97776.1 ABJB010531888 ABJB010904798 ABJB010920537 ABJB010953177 DS861388 EEC14104.1 GANO01002327 JAB57544.1 AJVK01022796 HACG01031276 CEK78141.1 GG666558 EEN55537.1 GBXI01006370 JAD07922.1 GDHF01018046 JAI34268.1 GDIP01016840 JAM86875.1 APGK01043395 BT128373 KB741014 KB632204 AEE63331.1 ENN75458.1 ERL90104.1 MG546685 AYB71126.1 NEDP02002833 OWF50001.1 CVRI01000055 CRL01100.1 RCHS01001867 RMX50980.1 ABJB010645197 DS674754 EEC03853.1 AAMC01008482 GBZX01000125 JAG92615.1 GDIQ01087961 JAN06776.1 GL193954 EFB23139.1 AAGW02063829 AANG04002472
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000027135
UP000015103
+ More
UP000009046 UP000235965 UP000007151 UP000215335 UP000007266 UP000075809 UP000000311 UP000053825 UP000053240 UP000053097 UP000279307 UP000242457 UP000005205 UP000002358 UP000078541 UP000007755 UP000008237 UP000078542 UP000078492 UP000054359 UP000005203 UP000192223 UP000053105 UP000245037 UP000002320 UP000008820 UP000094527 UP000092461 UP000069940 UP000249989 UP000245119 UP000079169 UP000076502 UP000000305 UP000076858 UP000030746 UP000001555 UP000092462 UP000001554 UP000076420 UP000085678 UP000019118 UP000030742 UP000242188 UP000183832 UP000275408 UP000007646 UP000008143 UP000001811 UP000286642 UP000011712
UP000009046 UP000235965 UP000007151 UP000215335 UP000007266 UP000075809 UP000000311 UP000053825 UP000053240 UP000053097 UP000279307 UP000242457 UP000005205 UP000002358 UP000078541 UP000007755 UP000008237 UP000078542 UP000078492 UP000054359 UP000005203 UP000192223 UP000053105 UP000245037 UP000002320 UP000008820 UP000094527 UP000092461 UP000069940 UP000249989 UP000245119 UP000079169 UP000076502 UP000000305 UP000076858 UP000030746 UP000001555 UP000092462 UP000001554 UP000076420 UP000085678 UP000019118 UP000030742 UP000242188 UP000183832 UP000275408 UP000007646 UP000008143 UP000001811 UP000286642 UP000011712
Interpro
SUPFAM
SSF47090
SSF47090
Gene 3D
ProteinModelPortal
H9JB94
A0A2A4J7B0
A0A2H1X361
A0A1E1WTB3
A0A0L7L2Z5
A0A194PE48
+ More
S4Q007 A0A2R7X5N5 A0A0A9YIW4 A0A067QKD5 A0A023FAM5 A0A224XVF6 R4G3H4 E0VMW4 A0A1B6DUX0 A0A2J7RBF7 A0A212ERP5 A0A232FAP1 D6X2Z1 A0A151WT91 E2AHG0 A0A0L7R1B2 A0A194QKS9 A0A026WDQ9 A0A2A3EHG7 A0A158P1Y9 A0A0C9RJU1 K7IQ78 A0A195FIF8 A0A1Y1KYB5 F4W665 E2BED3 A0A151IKI3 A0A195DMK1 T1IK58 A0A087UEZ7 A0A0T6B0S6 A0A087ZPN7 A0A1W4XC72 A0A0M9A408 A0A336LAH7 A0A336LE26 A0A2P8ZIZ5 A0A023FP57 A0A293N757 A0A0N7ZC60 A0A0K2TR34 A0A023FY87 G3MT06 B0W522 A0A023GP25 A0A1Q3FQN9 C1C2E4 Q17BA6 L7M877 A0A1D2N7P4 C1BRJ2 A0A023EPQ3 A0A1B0C927 A0A182H108 A0A2T7PDX7 A0A1S3D6Z9 A0A154NY41 A0A310SKQ4 E9H405 A0A0P5P9U2 A0A0P6HQS2 A0A131YPC0 A0A0P5ZMB7 A0A0P5CJL4 A0A1L8DLR5 A0A1L8DJS8 V5H0M4 A0A131YAK0 V5I1P0 V4AVB5 B7Q5I2 U5EJ84 A0A1B0D2G8 A0A0B7ADM8 C3YVX3 A0A0A1X9Z8 A0A0K8V6C0 A0A0P6BVP7 A0A2C9JGE2 A0A1S3IK20 J3JYP3 A0A385UIK0 A0A210QMS2 A0A1J1ING9 A0A3M6UBE1 G3TDG2 B7PB81 F7CD34 A0A0C9R6N9 A0A0P6CTJ9 D2I156 G1U070 A0A3Q7Y2P9 M3VYF8
S4Q007 A0A2R7X5N5 A0A0A9YIW4 A0A067QKD5 A0A023FAM5 A0A224XVF6 R4G3H4 E0VMW4 A0A1B6DUX0 A0A2J7RBF7 A0A212ERP5 A0A232FAP1 D6X2Z1 A0A151WT91 E2AHG0 A0A0L7R1B2 A0A194QKS9 A0A026WDQ9 A0A2A3EHG7 A0A158P1Y9 A0A0C9RJU1 K7IQ78 A0A195FIF8 A0A1Y1KYB5 F4W665 E2BED3 A0A151IKI3 A0A195DMK1 T1IK58 A0A087UEZ7 A0A0T6B0S6 A0A087ZPN7 A0A1W4XC72 A0A0M9A408 A0A336LAH7 A0A336LE26 A0A2P8ZIZ5 A0A023FP57 A0A293N757 A0A0N7ZC60 A0A0K2TR34 A0A023FY87 G3MT06 B0W522 A0A023GP25 A0A1Q3FQN9 C1C2E4 Q17BA6 L7M877 A0A1D2N7P4 C1BRJ2 A0A023EPQ3 A0A1B0C927 A0A182H108 A0A2T7PDX7 A0A1S3D6Z9 A0A154NY41 A0A310SKQ4 E9H405 A0A0P5P9U2 A0A0P6HQS2 A0A131YPC0 A0A0P5ZMB7 A0A0P5CJL4 A0A1L8DLR5 A0A1L8DJS8 V5H0M4 A0A131YAK0 V5I1P0 V4AVB5 B7Q5I2 U5EJ84 A0A1B0D2G8 A0A0B7ADM8 C3YVX3 A0A0A1X9Z8 A0A0K8V6C0 A0A0P6BVP7 A0A2C9JGE2 A0A1S3IK20 J3JYP3 A0A385UIK0 A0A210QMS2 A0A1J1ING9 A0A3M6UBE1 G3TDG2 B7PB81 F7CD34 A0A0C9R6N9 A0A0P6CTJ9 D2I156 G1U070 A0A3Q7Y2P9 M3VYF8
PDB
2KR2
E-value=3.43357e-58,
Score=568
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.948966
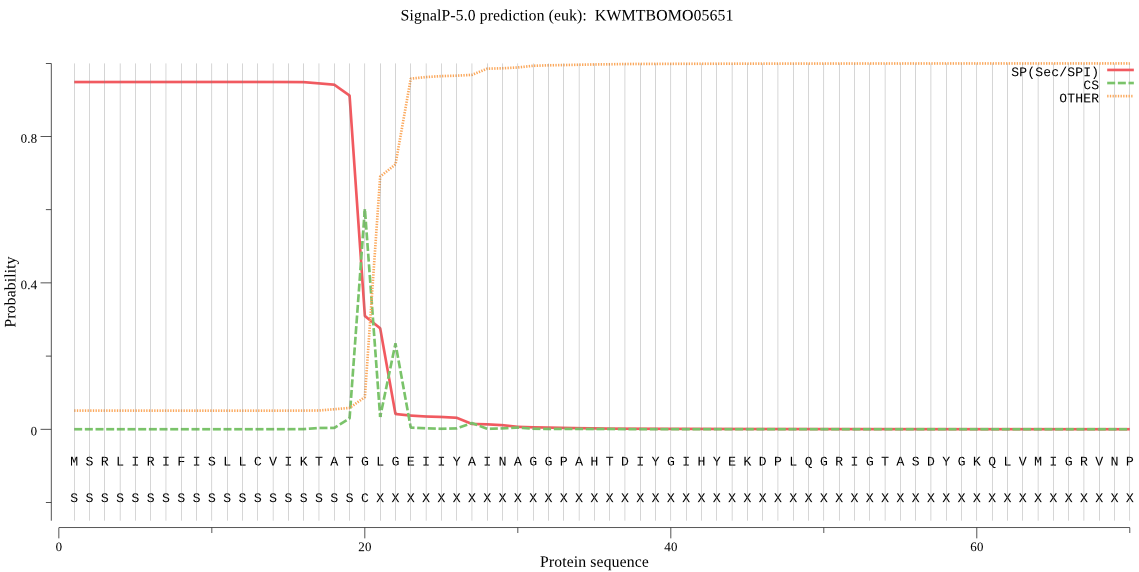
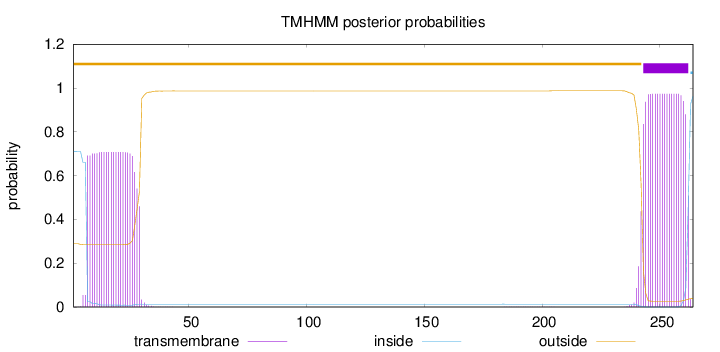
Length:
264
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.37185
Exp number, first 60 AAs:
15.87865
Total prob of N-in:
0.71118
POSSIBLE N-term signal
sequence
outside
1 - 242
TMhelix
243 - 262
inside
263 - 264
Population Genetic Test Statistics
Pi
218.676838
Theta
208.081159
Tajima's D
0.139717
CLR
54.370008
CSRT
0.410479476026199
Interpretation
Uncertain
