Gene
KWMTBOMO05650
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.853
Sequence
CDS
ATGTCGTCAGGTAGTTATTTATTAAATGTGCCGAAATTAAAAGGACGCGAAAATTATGACGACTGGGCATTCGCAGCAAGAAACTTTCTTATCTTGGAAGGCATAGACATCGACGCCATTCCGGTTGATTTGAGTCATGCCGAAGACCGAAAAGCGAAAGCAAAACTCGTCATGACCATTGATCCCAAGCTATATGTGCACATCAAAAATGAAACCAAAGTCAAGGATTTGTGGCAGAAACTACAAAAGTTATTCGATGACAGCGGGTTCACAAGAAAAATCAGCCTACTTCGAACACTGATATCCATTCGACTTGATAATTGTGAATCCATGACAAGTTACGTAAGTCAGATAGTGGAAACTGCACAAAGACTAAAAGGTACAGGGTTTGAAATTAATGAGGAATGGATCGGATCATTGCTGTTGGCGGGGCTGCCAGAAAAGTTTGCCCCGATGATTATGGCAATAGAACATTCCGGAATTGCCATAAATTCTGATGTCATAAAAACCAAATTGCTTGATATGAGTTCTGAGGTAGGTGACCATGACTCAGAAATTGCATTATGGTCGAAACAAAAACAAAGGGTAGAACAAAATTTTAATAAAAAATTTGAAAAACCAACTTCAAATTACACGCAAAATGTAAAAAAGATTATAAAATGCTACAAGTGCAAGAAAATTGGCCATTTTAAGAATCAGTGTCCTGAATTGAAAGAAAAGCAAGTAAACGCATTTAGTGCTGCTTTTTTTCATAGTGAATTTAATAGGGAAGACTGGTATATCGACTCAGGGGCTAGTGCTCATATGACATCAAGCAAAGAAAACTTATTGCAAGTGAGAAAAGTACACGATATGAAGGAAATTATTGTTGCAAATAATATGACTGTGCCAGTAGTGTGTGCCGGAAATTCACAGATTACTACTGTTGTGAATAATTCTCAGTTTGATATTAAGGTAAACAACATTTTATACATACCTAATCTAACAACAAACTTATTGTCAGTAAGCCAGTTGATAGCAAGAGGAAACAAAGTTTCTTTCAAAAGTGATGTATGTTATATACATAATCAACGAAATGAGCTTATTGGAATAGCAAATCTACAAAATGGTGTGTATAAGTTGAATACAGTAAGATCAGAGAAAGTGTTGGCGGCAGCAGTTCAGACTACAGATGCAAAACAGTGGCACAGTAGATTGGGACACATTAATAGTAATGATTTGCAGAAGATGAAAAATGGTGCTGTAGACGGAATATCGTTTGATATGAAGGCTAATATAAAAAAATCCAACTGTCAAGTATGTTGTGAAGGGAAACAGAGTCGTTTACCGTTCCAATTACGAAACCATAGAAGTACAGAATTGCTAGATTTAGTTCACTCAGATCTGTGTGGGCCTATGGAGGTTCGATCGATTGGACAGGCAAGATATTTTCTATTGTTTGTTGACGATGCTAGCAGGATGGCTTTTGTATATTTTATAAAGGAGAAGAATCAGACTCTACAGCGTTTCAAGGAATTTCAAAAAATGGTTGAGAATCAAAAAGGAACAAAGATAAAAACATTGAGAACCGACAATGGAGGGGAGTTTTCTTCAGATATGATGGAAAATTATCTGATAGATAGTGGAATTATCCATCAGAAGACGAACCCATATACTCCGGAGCAGAATGGTCTCAGCGAACGTTTCAACAGGACTATAGTTGAACGTGCAAGATGTTTATTGTTCGAGGCCAAACTGGATAAAAGTTTTTGGGCAGAAGCAGTTAACACGGCAGTATACTTGAAAAATAGATCACCGGCTTCAGGTTTGGACCAAAAGACACCGATTGAAGTATGGACAGGACAAAAACCTGATCTCAGTCATGTCAGAATATTTGGGAGCCCAGTAATGATGCATGTTCCTAAAAATAAAAGATTGAAATGGGACAAGAAAGCAGTTCAATTCATATTAGTGGGGTATCCAGAGAATGTGAAAGGCTACAGATTGTATAACTTAGAAACTAGAAAAATATGTACCAGCAGAGATGTCATTGTAATGGAACCAGATACTACTCAAATAGAAATAAACGAAGGACAGATAGAAAAACAGCAGTTAGCACAAGAGGAAAGTTCTACTGAAGAAAGTGAAATAACATTGACAAATCAGGATGATCCCACTTATGTATATGATTCAGAAAAAAGTACAACTTCTACTGAAGGAGACTCTTATCAGACATTATCTGAAGAAGAAGAGCTATCTGAGAAAGAAGAGCCTCGATTGGTGGAAAAGCGTACCAGGAAACCTCCTGAAAGGTATGGTTTTACCAATCTTTGTATTTCAGATGAACCAGAGTCTCGGTTTGAAGACATAACTTATGATGAAGCTACCAACGGTCCAGAGAAAGACAAGTGGCAGCAGGCTATGTCTGATGAACTTCAAGCGTTTGAAGACAACCAAGCGTGGGAGATTGTTAAACAAGAGGAGGCAGATAGAGTAGTACAGTGCAAATGGGTGTTCAAGAAAAAGCTAGAAAGCAATAACAGTGTTAGGTATAGAGCTAGACTTGTAGCGAAAGGTTACACTCAGCAGCATGGAGTAGATTTTAACGAAACTTACTCACCAGTGCTTAGATACTCTACACTTAGACTTCTGTTTGCATTAAGTGTAAAATTTCATTTTGAAATCTTTCATCTTGATGTAGTTACAGCATTTTTAAATGGTTTTTTAACAGAAAATGTGTACATGTACGCACCTCCTAGCTTAAATTGTCCAAGTAATTGTGTTCTTAAACTTAAAAAAGCAGTTTATGGTTTGAAGCAGTCTGCTAGGGCATGGAATACCAGAATAAATGATTATTTAATAGACATGGATTATGTAAAGTCAGTTTCAGAACCATGTTTGTATAGTAAAAAATGTAAAAATGAGCAGATTATCATTGCAATATTTGTTGATGATTTTTTCATTTTTTCTAATAGTAAGACTATGACTAATGATTTCAAGAAAAAGCTAATGTCAAAATTTTAA
Protein
MSSGSYLLNVPKLKGRENYDDWAFAARNFLILEGIDIDAIPVDLSHAEDRKAKAKLVMTIDPKLYVHIKNETKVKDLWQKLQKLFDDSGFTRKISLLRTLISIRLDNCESMTSYVSQIVETAQRLKGTGFEINEEWIGSLLLAGLPEKFAPMIMAIEHSGIAINSDVIKTKLLDMSSEVGDHDSEIALWSKQKQRVEQNFNKKFEKPTSNYTQNVKKIIKCYKCKKIGHFKNQCPELKEKQVNAFSAAFFHSEFNREDWYIDSGASAHMTSSKENLLQVRKVHDMKEIIVANNMTVPVVCAGNSQITTVVNNSQFDIKVNNILYIPNLTTNLLSVSQLIARGNKVSFKSDVCYIHNQRNELIGIANLQNGVYKLNTVRSEKVLAAAVQTTDAKQWHSRLGHINSNDLQKMKNGAVDGISFDMKANIKKSNCQVCCEGKQSRLPFQLRNHRSTELLDLVHSDLCGPMEVRSIGQARYFLLFVDDASRMAFVYFIKEKNQTLQRFKEFQKMVENQKGTKIKTLRTDNGGEFSSDMMENYLIDSGIIHQKTNPYTPEQNGLSERFNRTIVERARCLLFEAKLDKSFWAEAVNTAVYLKNRSPASGLDQKTPIEVWTGQKPDLSHVRIFGSPVMMHVPKNKRLKWDKKAVQFILVGYPENVKGYRLYNLETRKICTSRDVIVMEPDTTQIEINEGQIEKQQLAQEESSTEESEITLTNQDDPTYVYDSEKSTTSTEGDSYQTLSEEEELSEKEEPRLVEKRTRKPPERYGFTNLCISDEPESRFEDITYDEATNGPEKDKWQQAMSDELQAFEDNQAWEIVKQEEADRVVQCKWVFKKKLESNNSVRYRARLVAKGYTQQHGVDFNETYSPVLRYSTLRLLFALSVKFHFEIFHLDVVTAFLNGFLTENVYMYAPPSLNCPSNCVLKLKKAVYGLKQSARAWNTRINDYLIDMDYVKSVSEPCLYSKKCKNEQIIIAIFVDDFFIFSNSKTMTNDFKKKLMSKF
Summary
Uniprot
A0A1Y1N668
A0A212F7V4
A0A1Y1N932
V5GAG2
A0A194RJV9
A0A0J7NHJ8
+ More
A0A194RU38 A0A1Y1N6L8 A0A1Y1NAZ7 A0A2A4J959 A0A2A4JRI1 A0A1Y1N664 A0A0A1WXQ3 A0A0N1ICY1 A0A034VP70 A0A1W7R6K6 A0A1W7R6J6 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2J7R668 A0A085MVP5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V1APS7 A0A355BEE2 A0A0V0TUP0 A0A085N572 A0A0A9WQD4 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A1Y1LYU4 A0A1Y1N3X2 K7J8U2 A0A0K8U6S2 A0A0J7KG03 A0A0A1XLZ7 A0A0V0Z6Q0 A0A085MSF4 A0A182GZN1 A0A0A1WFC7 A0A0J7K6W1 A0A0V0YNF4 A0A0V0RWJ3 A0A085N9K0 A0A085NQR3 A0A1Y1LP39 A0A224X581 A0A0V1APU2 A0A2M3YZL7 A0A2M3YZL3 A0A2M4CV35 A0A1W7R6H7 A0A0A1X768 W8AQ86 A0A1Y1MYL6 A0A1Y1MYL8 A0A0V1GS48 A0A0J7K7P3 W8B7K5 A0A0J7K7B7 A0A0K8U956 A0A0V1MGJ0 A0A0V1NJN8 A0A2N9IWX1 A0A251V331 Q9ZQE9 B1N668 Q6L3N8 A0A0V1GTY0 A0A0V0V098 A0A3S3NIE3 A0A0V0IV83 A0A1W7R6I5 A0A2J7PSB2 A0A2N9FSJ8 A0A151R7U0 A0A068B703 A0A131XNF9 A0A396GW39 A0A2N9ETM3 A0A0J7K9H9
A0A194RU38 A0A1Y1N6L8 A0A1Y1NAZ7 A0A2A4J959 A0A2A4JRI1 A0A1Y1N664 A0A0A1WXQ3 A0A0N1ICY1 A0A034VP70 A0A1W7R6K6 A0A1W7R6J6 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2J7R668 A0A085MVP5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V1APS7 A0A355BEE2 A0A0V0TUP0 A0A085N572 A0A0A9WQD4 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A1Y1LYU4 A0A1Y1N3X2 K7J8U2 A0A0K8U6S2 A0A0J7KG03 A0A0A1XLZ7 A0A0V0Z6Q0 A0A085MSF4 A0A182GZN1 A0A0A1WFC7 A0A0J7K6W1 A0A0V0YNF4 A0A0V0RWJ3 A0A085N9K0 A0A085NQR3 A0A1Y1LP39 A0A224X581 A0A0V1APU2 A0A2M3YZL7 A0A2M3YZL3 A0A2M4CV35 A0A1W7R6H7 A0A0A1X768 W8AQ86 A0A1Y1MYL6 A0A1Y1MYL8 A0A0V1GS48 A0A0J7K7P3 W8B7K5 A0A0J7K7B7 A0A0K8U956 A0A0V1MGJ0 A0A0V1NJN8 A0A2N9IWX1 A0A251V331 Q9ZQE9 B1N668 Q6L3N8 A0A0V1GTY0 A0A0V0V098 A0A3S3NIE3 A0A0V0IV83 A0A1W7R6I5 A0A2J7PSB2 A0A2N9FSJ8 A0A151R7U0 A0A068B703 A0A131XNF9 A0A396GW39 A0A2N9ETM3 A0A0J7K9H9
Pubmed
EMBL
GEZM01011856
JAV93393.1
AGBW02009826
OWR49812.1
GEZM01011857
JAV93390.1
+ More
GALX01001351 JAB67115.1 KQ460297 KPJ16236.1 LBMM01004924 KMQ91985.1 KQ459875 KPJ19636.1 GEZM01011858 GEZM01011854 JAV93389.1 GEZM01011855 JAV93396.1 NWSH01002292 PCG68657.1 PCG68658.1 NWSH01000816 PCG74073.1 GEZM01011859 JAV93384.1 GBXI01011094 JAD03198.1 KQ460882 KPJ11482.1 GAKP01015352 JAC43600.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 NEVH01019375 PNF22597.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 NEVH01006980 PNF36330.1 KL367628 KFD61291.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 JYDJ01000808 KRX33413.1 JYDH01000330 KRY26802.1 DNYX01000353 HBK84078.1 JYDJ01000136 KRX42725.1 KL367553 KFD64618.1 GBHO01036544 JAG07060.1 GBHO01024949 JAG18655.1 GBHO01024951 JAG18653.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 GEZM01045163 GEZM01045162 JAV77888.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 AAZX01000499 GDHF01030254 JAI22060.1 LBMM01008124 KMQ89126.1 GBXI01002679 JAD11613.1 JYDQ01000379 KRY07987.1 KL367691 KFD60150.1 JXUM01099923 KQ564528 KXJ72133.1 GBXI01017107 JAC97184.1 LBMM01012645 KMQ86082.1 JYDU01000001 KRY01876.1 JYDL01000068 KRX18751.1 KL367527 KFD66146.1 KL367480 KFD71809.1 GEZM01050757 JAV75412.1 GFTR01008771 JAW07655.1 KRY26801.1 GGFM01000952 MBW21703.1 GGFM01000953 MBW21704.1 GGFL01004951 MBW69129.1 GEHC01000879 JAV46766.1 GBXI01007143 JAD07149.1 GAMC01018338 JAB88217.1 GEZM01017329 JAV90762.1 GEZM01017328 JAV90763.1 JYDP01000350 KRZ01021.1 LBMM01012464 KMQ86211.1 GAMC01020736 JAB85819.1 LBMM01012669 KMQ86061.1 GDHF01029191 JAI23123.1 JYDO01000104 KRZ71003.1 JYDM01000185 KRZ84108.1 OIVN01006239 SPD28653.1 CM007893 OTG30017.1 AC006248 AAD17409.1 EF094939 EF094940 ABO36622.1 ABO36636.1 AC149291 AAT38758.1 JYDP01000258 KRZ01816.1 JYDN01000113 KRX56900.1 NCKU01012701 RWS00004.1 GEDG01001927 JAP36462.1 GEHC01000878 JAV46767.1 NEVH01021933 PNF19227.1 OIVN01001130 SPC90232.1 KQ483989 KYP38573.1 KF740825 AIC77183.1 GEFH01000951 JAP67630.1 PSQE01000007 RHN45312.1 OIVN01000313 SPC78143.1 LBMM01011374 KMQ86881.1
GALX01001351 JAB67115.1 KQ460297 KPJ16236.1 LBMM01004924 KMQ91985.1 KQ459875 KPJ19636.1 GEZM01011858 GEZM01011854 JAV93389.1 GEZM01011855 JAV93396.1 NWSH01002292 PCG68657.1 PCG68658.1 NWSH01000816 PCG74073.1 GEZM01011859 JAV93384.1 GBXI01011094 JAD03198.1 KQ460882 KPJ11482.1 GAKP01015352 JAC43600.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 NEVH01019375 PNF22597.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 NEVH01006980 PNF36330.1 KL367628 KFD61291.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 JYDR01000001 KRY80095.1 JYDJ01000808 KRX33413.1 JYDH01000330 KRY26802.1 DNYX01000353 HBK84078.1 JYDJ01000136 KRX42725.1 KL367553 KFD64618.1 GBHO01036544 JAG07060.1 GBHO01024949 JAG18655.1 GBHO01024951 JAG18653.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 GEZM01045163 GEZM01045162 JAV77888.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 AAZX01000499 GDHF01030254 JAI22060.1 LBMM01008124 KMQ89126.1 GBXI01002679 JAD11613.1 JYDQ01000379 KRY07987.1 KL367691 KFD60150.1 JXUM01099923 KQ564528 KXJ72133.1 GBXI01017107 JAC97184.1 LBMM01012645 KMQ86082.1 JYDU01000001 KRY01876.1 JYDL01000068 KRX18751.1 KL367527 KFD66146.1 KL367480 KFD71809.1 GEZM01050757 JAV75412.1 GFTR01008771 JAW07655.1 KRY26801.1 GGFM01000952 MBW21703.1 GGFM01000953 MBW21704.1 GGFL01004951 MBW69129.1 GEHC01000879 JAV46766.1 GBXI01007143 JAD07149.1 GAMC01018338 JAB88217.1 GEZM01017329 JAV90762.1 GEZM01017328 JAV90763.1 JYDP01000350 KRZ01021.1 LBMM01012464 KMQ86211.1 GAMC01020736 JAB85819.1 LBMM01012669 KMQ86061.1 GDHF01029191 JAI23123.1 JYDO01000104 KRZ71003.1 JYDM01000185 KRZ84108.1 OIVN01006239 SPD28653.1 CM007893 OTG30017.1 AC006248 AAD17409.1 EF094939 EF094940 ABO36622.1 ABO36636.1 AC149291 AAT38758.1 JYDP01000258 KRZ01816.1 JYDN01000113 KRX56900.1 NCKU01012701 RWS00004.1 GEDG01001927 JAP36462.1 GEHC01000878 JAV46767.1 NEVH01021933 PNF19227.1 OIVN01001130 SPC90232.1 KQ483989 KYP38573.1 KF740825 AIC77183.1 GEFH01000951 JAP67630.1 PSQE01000007 RHN45312.1 OIVN01000313 SPC78143.1 LBMM01011374 KMQ86881.1
Proteomes
UP000007151
UP000053240
UP000036403
UP000218220
UP000235965
UP000054995
+ More
UP000054805 UP000054826 UP000054632 UP000055048 UP000054776 UP000002358 UP000054783 UP000069940 UP000249989 UP000054815 UP000054630 UP000055024 UP000054843 UP000054924 UP000215914 UP000054681 UP000285301 UP000075243 UP000265566
UP000054805 UP000054826 UP000054632 UP000055048 UP000054776 UP000002358 UP000054783 UP000069940 UP000249989 UP000054815 UP000054630 UP000055024 UP000054843 UP000054924 UP000215914 UP000054681 UP000285301 UP000075243 UP000265566
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A1Y1N668
A0A212F7V4
A0A1Y1N932
V5GAG2
A0A194RJV9
A0A0J7NHJ8
+ More
A0A194RU38 A0A1Y1N6L8 A0A1Y1NAZ7 A0A2A4J959 A0A2A4JRI1 A0A1Y1N664 A0A0A1WXQ3 A0A0N1ICY1 A0A034VP70 A0A1W7R6K6 A0A1W7R6J6 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2J7R668 A0A085MVP5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V1APS7 A0A355BEE2 A0A0V0TUP0 A0A085N572 A0A0A9WQD4 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A1Y1LYU4 A0A1Y1N3X2 K7J8U2 A0A0K8U6S2 A0A0J7KG03 A0A0A1XLZ7 A0A0V0Z6Q0 A0A085MSF4 A0A182GZN1 A0A0A1WFC7 A0A0J7K6W1 A0A0V0YNF4 A0A0V0RWJ3 A0A085N9K0 A0A085NQR3 A0A1Y1LP39 A0A224X581 A0A0V1APU2 A0A2M3YZL7 A0A2M3YZL3 A0A2M4CV35 A0A1W7R6H7 A0A0A1X768 W8AQ86 A0A1Y1MYL6 A0A1Y1MYL8 A0A0V1GS48 A0A0J7K7P3 W8B7K5 A0A0J7K7B7 A0A0K8U956 A0A0V1MGJ0 A0A0V1NJN8 A0A2N9IWX1 A0A251V331 Q9ZQE9 B1N668 Q6L3N8 A0A0V1GTY0 A0A0V0V098 A0A3S3NIE3 A0A0V0IV83 A0A1W7R6I5 A0A2J7PSB2 A0A2N9FSJ8 A0A151R7U0 A0A068B703 A0A131XNF9 A0A396GW39 A0A2N9ETM3 A0A0J7K9H9
A0A194RU38 A0A1Y1N6L8 A0A1Y1NAZ7 A0A2A4J959 A0A2A4JRI1 A0A1Y1N664 A0A0A1WXQ3 A0A0N1ICY1 A0A034VP70 A0A1W7R6K6 A0A1W7R6J6 A0A2J7Q1Z2 A0A2J7RKJ6 A0A2J7R8S8 A0A2J7R668 A0A085MVP5 A0A0V1FKH2 A0A0V1K1Y9 A0A0V1F2E6 A0A0V0T3I0 A0A0V1APS7 A0A355BEE2 A0A0V0TUP0 A0A085N572 A0A0A9WQD4 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A1Y1LYU4 A0A1Y1N3X2 K7J8U2 A0A0K8U6S2 A0A0J7KG03 A0A0A1XLZ7 A0A0V0Z6Q0 A0A085MSF4 A0A182GZN1 A0A0A1WFC7 A0A0J7K6W1 A0A0V0YNF4 A0A0V0RWJ3 A0A085N9K0 A0A085NQR3 A0A1Y1LP39 A0A224X581 A0A0V1APU2 A0A2M3YZL7 A0A2M3YZL3 A0A2M4CV35 A0A1W7R6H7 A0A0A1X768 W8AQ86 A0A1Y1MYL6 A0A1Y1MYL8 A0A0V1GS48 A0A0J7K7P3 W8B7K5 A0A0J7K7B7 A0A0K8U956 A0A0V1MGJ0 A0A0V1NJN8 A0A2N9IWX1 A0A251V331 Q9ZQE9 B1N668 Q6L3N8 A0A0V1GTY0 A0A0V0V098 A0A3S3NIE3 A0A0V0IV83 A0A1W7R6I5 A0A2J7PSB2 A0A2N9FSJ8 A0A151R7U0 A0A068B703 A0A131XNF9 A0A396GW39 A0A2N9ETM3 A0A0J7K9H9
PDB
1C1A
E-value=0.00189348,
Score=102
Ontologies
KEGG
GO
PANTHER
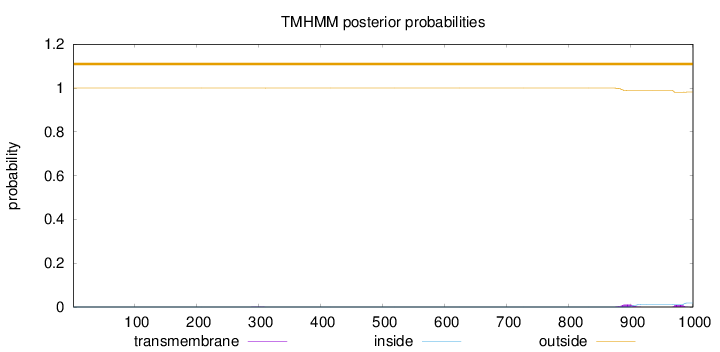
Topology
Length:
1000
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.3842
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.00025
outside
1 - 1000
Population Genetic Test Statistics
Pi
39.860176
Theta
142.176119
Tajima's D
-1.325357
CLR
139.542917
CSRT
0.0858957052147393
Interpretation
Uncertain
