Gene
KWMTBOMO05646
Pre Gene Modal
BGIBMGA006545
Annotation
kinesin-like_protein_Ncd_[Bombyx_mori]
Full name
Kinesin-like protein
+ More
Protein claret segregational
Protein claret segregational
Location in the cell
Nuclear Reliability : 4.611
Sequence
CDS
ATGTCCAAGATACCAAAACTACCAACAATATCAAAAGAAAATAGATTTGGACAATTTCATAATCGGCCCATATCCAGAACTATTGCTAATGGTCTCAGTGATGTTGATAAGAAAAATTTAATCACTAATCATACTAGACCTTTACGAAATGGCCCACCTGTTTCTGCTGCTGCACCTCGGATTAAGAGATCTGCAACTGCACCATCATCGACAACAATTCCGAATAATGTGACAAAAGTTGAAAAAAGATCAACTGTGGCTCCGAAAATCCCTCCTTATGACTTCAAAGCAAGATTCAATGACTTGTTAGAAAAACATAAAAAAATGAAGAGTGAGTTTACTGATTTAAAAGACAAACATTTAGAAGTCTCAGATGAATATGAAAAGATCAAAGAGACATTCCAAAGCTGTAGCAATGAAAGAGATATTCTTAAGGCCAATTTAAGTGTTAAAACTTTAGAGTATGATGAAATTAAAGTTAATTTTGATGTAATGAAAACAGAAAATGAAGATCTACAACGTAAGACAAAGCTTCTAGAGGAAGTTACATTTAGTCTTAAACAGAAATCTTTTGAACTAGATCAAGTTCAGACAGATTTAAATTCTTTAAGAAGACGCCACAGCAGCCTACAAGAAGAAGCAGAAGCACTAAGAGTACTCACTGACCAACTAAAAAAAAAATGCATTGAATATGACAAACTAAAATTAGATTTTACTGAAGCTCAGGAAAATATTATCAAATATAAAACTGATTCTGAAGCTCTCCAAAATATTTTGGCATCAATGTATAAAGAGCAGAGAGACCTTAGGAATGCTATTCAAGATTTAAAGGGTAACATTCGTGTATACTGCCGAATTAGACCACCTCTAAGTATTGAAATAACAAAGCCACTGTTTAATCTAAATGTTGTTGATGCTTGTTCTATAGAAGTTGAAAAAATAGAATTACTTAATTCAGCCAGAAAAACAAAACCACAATTATTTACTTTTGATGGCATATTTACTCCACATGCATCTCAGGAAGATGTTTTTGCAGAAGTTTCGTCAATGGTCCAATCGGCTTTAGATGGGTATAATGTGTGCATATTTGCTTATGGACAGACAGGATCAGGCAAGACATATACAATGGAAGGAGGTTGTGGAACCGAGCAGTATGGTATTATACCACGAGCCTTTAACATGATTTTTACATGCATGGAAGATTTGAAAAGAATGGGCTGGGAACTTACAATAAAAGCTTCATTTTTAGAGATTTACAATGAAGTCATATATGATTTACTCAATTCCAGTAAGGACCAGGAAAATCATGAAATTAAAATGGTTAATTCAAAGGGTGGTGATGTATATGTGTCAAACTTGAAAGAAGAAGAAGTCAAAAGTTCACACGAATTCATCAGGCTGATGATATTTGCCCAAAGAAACAGACAAACTGCTGCTACATTGAACAATGAACGCAGCTCCAGATCTCATTCGGTGGCTCAAATTAAAATAGCTGCTATAAATGAGAAGCGAAAGGAAAAGTACACTAGTAATTTAAATCTAGTTGACTTAGCTGGGTCTGAAAGTGGAAAGACAACACAAAGAATGGATGAAACTAAACATATCAATAGGTCTTTGTCTGAATTATCTAAAGTTATTCTTTCCCTCCAAACAAATCAGATGCATATTCCTTACAGAAATTCCAAATTGACTCACTTATTAATGCCTAGTCTTGGTGGCAACTCTAAGACACTTATGTTGGTCAATATCAATCAATTTGATGAGTGCTTTAGTGAAACTTTGAATTCGTTGAGGTTTGCTACAAAAGTCAATAACTGTAGAGTAGTTAAGGCCAAGAAGAACCTATGTATGGTCGACACTTAG
Protein
MSKIPKLPTISKENRFGQFHNRPISRTIANGLSDVDKKNLITNHTRPLRNGPPVSAAAPRIKRSATAPSSTTIPNNVTKVEKRSTVAPKIPPYDFKARFNDLLEKHKKMKSEFTDLKDKHLEVSDEYEKIKETFQSCSNERDILKANLSVKTLEYDEIKVNFDVMKTENEDLQRKTKLLEEVTFSLKQKSFELDQVQTDLNSLRRRHSSLQEEAEALRVLTDQLKKKCIEYDKLKLDFTEAQENIIKYKTDSEALQNILASMYKEQRDLRNAIQDLKGNIRVYCRIRPPLSIEITKPLFNLNVVDACSIEVEKIELLNSARKTKPQLFTFDGIFTPHASQEDVFAEVSSMVQSALDGYNVCIFAYGQTGSGKTYTMEGGCGTEQYGIIPRAFNMIFTCMEDLKRMGWELTIKASFLEIYNEVIYDLLNSSKDQENHEIKMVNSKGGDVYVSNLKEEEVKSSHEFIRLMIFAQRNRQTAATLNNERSSRSHSVAQIKIAAINEKRKEKYTSNLNLVDLAGSESGKTTQRMDETKHINRSLSELSKVILSLQTNQMHIPYRNSKLTHLLMPSLGGNSKTLMLVNINQFDECFSETLNSLRFATKVNNCRVVKAKKNLCMVDT
Summary
Description
NCD is required for normal chromosomal segregation in meiosis, in females, and in early mitotic divisions of the embryo. The NCD motor activity is directed toward the microtubule's minus end.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. NCD subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. NCD subfamily.
Keywords
3D-structure
ATP-binding
Cell cycle
Cell division
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Meiosis
Microtubule
Mitosis
Motor protein
Nucleotide-binding
Phosphoprotein
Reference proteome
Feature
chain Protein claret segregational
Uniprot
H9JAK0
A1E130
A0A3S2LKH3
A0A2A4J703
A0A2H1WLZ7
A0A0L7LN46
+ More
A0A212F9G4 A0A194PE52 A0A194QKT3 A0A1Q3FEE7 A0A1Q3FEI2 A0A336LWZ5 B0X1T3 A0A182LQH0 A0A0M4F5S2 A0A182HZG2 A0A182X398 A0A182JT54 Q7PTK6 A0A182VN44 A0A182UGG8 B4PPJ6 A0A182G315 A0A084W2N5 A0A0L0BYK3 A0A1A9YQ61 B4G304 Q297T2 A0A1B0B2Z0 A0A0A1X047 Q16RX2 A0A154PQN6 A0A1A9ZSV7 B3LWX8 A0A1S4FRB8 A0A182NMP0 A0A1I8PCQ8 A0A0C9QWK4 A0A3B0JL46 A0A0K8U4R0 A0A182R079 B4HZF4 A0A182PCR1 A0A3B0K7S6 A0A034W445 B3P5C7 A0A1A9VDP2 A0A0B4LI25 P20480 A0A1W4UC82 W8C7M3 A0A088A154 A0A1B0FL90 A0A0L7R3V3 A0A182LWB3 A0A0L7QW86 A0A2A3ELK3 K7J2W4 B4M0Q0 A0A1A9W5P3 A0A182VQ27 B4NBM4 A0A182Y4Q2 B4JRJ6 A0A232F0M7 A0A195EVA0 A0A151X3M4 A0A182RH46 A0A1I8MBC3 A0A1J1HYB5 B4KAN3 A0A195E5Y2 A0A1B0DP57 A0A151HZ56 A0A158NR31 F4WPB2 E2C5K6 A0A151IN90 A0A182FSW8 W5J5H6 A0A182JKR6 A0A026WWH4 A0A3L8D998 A0A1B0EWA3 A0A1B0BZB5 E2AJ68 A0A1A9XDN0 A0A1B0G3E0 B6UXG3 B6UXD7 A0A0M9A0I4 B6UXE7 B6UXD2 A0A1A9UMT1 B6UXE3 B6UXG0 B6UXF5 B6UXE0 B6UXE4
A0A212F9G4 A0A194PE52 A0A194QKT3 A0A1Q3FEE7 A0A1Q3FEI2 A0A336LWZ5 B0X1T3 A0A182LQH0 A0A0M4F5S2 A0A182HZG2 A0A182X398 A0A182JT54 Q7PTK6 A0A182VN44 A0A182UGG8 B4PPJ6 A0A182G315 A0A084W2N5 A0A0L0BYK3 A0A1A9YQ61 B4G304 Q297T2 A0A1B0B2Z0 A0A0A1X047 Q16RX2 A0A154PQN6 A0A1A9ZSV7 B3LWX8 A0A1S4FRB8 A0A182NMP0 A0A1I8PCQ8 A0A0C9QWK4 A0A3B0JL46 A0A0K8U4R0 A0A182R079 B4HZF4 A0A182PCR1 A0A3B0K7S6 A0A034W445 B3P5C7 A0A1A9VDP2 A0A0B4LI25 P20480 A0A1W4UC82 W8C7M3 A0A088A154 A0A1B0FL90 A0A0L7R3V3 A0A182LWB3 A0A0L7QW86 A0A2A3ELK3 K7J2W4 B4M0Q0 A0A1A9W5P3 A0A182VQ27 B4NBM4 A0A182Y4Q2 B4JRJ6 A0A232F0M7 A0A195EVA0 A0A151X3M4 A0A182RH46 A0A1I8MBC3 A0A1J1HYB5 B4KAN3 A0A195E5Y2 A0A1B0DP57 A0A151HZ56 A0A158NR31 F4WPB2 E2C5K6 A0A151IN90 A0A182FSW8 W5J5H6 A0A182JKR6 A0A026WWH4 A0A3L8D998 A0A1B0EWA3 A0A1B0BZB5 E2AJ68 A0A1A9XDN0 A0A1B0G3E0 B6UXG3 B6UXD7 A0A0M9A0I4 B6UXE7 B6UXD2 A0A1A9UMT1 B6UXE3 B6UXG0 B6UXF5 B6UXE0 B6UXE4
Pubmed
19121390
20368730
26227816
22118469
26354079
20966253
+ More
12364791 14747013 17210077 17994087 17550304 26483478 24438588 26108605 15632085 25830018 17510324 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1691829 12537569 2140958 2146510 8112290 1825056 8670831 18327897 8606780 24495485 20075255 25244985 28648823 25315136 21347285 21719571 20798317 20920257 23761445 24508170 30249741 18984573
12364791 14747013 17210077 17994087 17550304 26483478 24438588 26108605 15632085 25830018 17510324 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1691829 12537569 2140958 2146510 8112290 1825056 8670831 18327897 8606780 24495485 20075255 25244985 28648823 25315136 21347285 21719571 20798317 20920257 23761445 24508170 30249741 18984573
EMBL
BABH01008799
EF092449
ABK92269.1
RSAL01000080
RVE48611.1
NWSH01002607
+ More
PCG67865.1 ODYU01009576 SOQ54101.1 JTDY01000483 KOB76973.1 AGBW02009576 OWR50395.1 KQ459606 KPI91542.1 KQ461198 KPJ06137.1 GFDL01009108 JAV25937.1 GFDL01009075 JAV25970.1 UFQS01000177 UFQT01000177 SSX00803.1 SSX21183.1 DS232269 EDS38811.1 CP012526 ALC46920.1 APCN01001975 AAAB01008799 EAA03777.5 CM000160 EDW98246.1 JXUM01140582 JXUM01140583 KQ569087 KXJ68769.1 ATLV01019658 KE525276 KFB44479.1 JRES01001160 KNC25056.1 CH479179 EDW24199.1 CM000070 EAL28123.2 JXJN01007738 GBXI01010269 JAD04023.1 CH477694 EAT37177.1 KQ435050 KZC14236.1 CH902617 EDV43817.1 GBYB01000095 JAG69862.1 OUUW01000005 SPP81052.1 GDHF01031029 JAI21285.1 AXCN02001212 CH480819 EDW53411.1 SPP81051.1 GAKP01008606 JAC50346.1 CH954182 EDV53177.1 AE014297 AHN57591.1 X52814 M33932 AY058596 X57475 GAMC01003536 GAMC01003534 JAC03022.1 CCAG010013281 KQ414663 KOC65509.1 AXCM01000279 KQ414716 KOC62824.1 KZ288215 PBC32683.1 CH940650 EDW67342.1 CH964232 EDW81188.1 CH916373 EDV94386.1 NNAY01001399 OXU24109.1 KQ981965 KYN31814.1 KQ982558 KYQ54991.1 CVRI01000036 CRK93080.1 CH933806 EDW16770.1 KQ979608 KYN20496.1 AJVK01007914 KQ976717 KYM76772.1 ADTU01023780 GL888243 EGI64093.1 GL452770 EFN76819.1 KQ976946 KYN06934.1 ADMH02002125 ETN58653.1 KK107078 EZA60402.1 QOIP01000011 RLU16712.1 AJWK01022514 JXJN01023088 GL439967 EFN66508.1 CCAG010009526 FJ219211 ACI96507.1 FJ219185 ACI96481.1 KQ435776 KOX74885.1 FJ219195 FJ219197 ACI96491.1 ACI96493.1 FJ219180 ACI96476.1 FJ219191 ACI96487.1 FJ219208 ACI96504.1 FJ219203 ACI96499.1 FJ219188 FJ219196 FJ219198 FJ219210 ACI96484.1 ACI96492.1 ACI96494.1 ACI96506.1 FJ219192 ACI96488.1
PCG67865.1 ODYU01009576 SOQ54101.1 JTDY01000483 KOB76973.1 AGBW02009576 OWR50395.1 KQ459606 KPI91542.1 KQ461198 KPJ06137.1 GFDL01009108 JAV25937.1 GFDL01009075 JAV25970.1 UFQS01000177 UFQT01000177 SSX00803.1 SSX21183.1 DS232269 EDS38811.1 CP012526 ALC46920.1 APCN01001975 AAAB01008799 EAA03777.5 CM000160 EDW98246.1 JXUM01140582 JXUM01140583 KQ569087 KXJ68769.1 ATLV01019658 KE525276 KFB44479.1 JRES01001160 KNC25056.1 CH479179 EDW24199.1 CM000070 EAL28123.2 JXJN01007738 GBXI01010269 JAD04023.1 CH477694 EAT37177.1 KQ435050 KZC14236.1 CH902617 EDV43817.1 GBYB01000095 JAG69862.1 OUUW01000005 SPP81052.1 GDHF01031029 JAI21285.1 AXCN02001212 CH480819 EDW53411.1 SPP81051.1 GAKP01008606 JAC50346.1 CH954182 EDV53177.1 AE014297 AHN57591.1 X52814 M33932 AY058596 X57475 GAMC01003536 GAMC01003534 JAC03022.1 CCAG010013281 KQ414663 KOC65509.1 AXCM01000279 KQ414716 KOC62824.1 KZ288215 PBC32683.1 CH940650 EDW67342.1 CH964232 EDW81188.1 CH916373 EDV94386.1 NNAY01001399 OXU24109.1 KQ981965 KYN31814.1 KQ982558 KYQ54991.1 CVRI01000036 CRK93080.1 CH933806 EDW16770.1 KQ979608 KYN20496.1 AJVK01007914 KQ976717 KYM76772.1 ADTU01023780 GL888243 EGI64093.1 GL452770 EFN76819.1 KQ976946 KYN06934.1 ADMH02002125 ETN58653.1 KK107078 EZA60402.1 QOIP01000011 RLU16712.1 AJWK01022514 JXJN01023088 GL439967 EFN66508.1 CCAG010009526 FJ219211 ACI96507.1 FJ219185 ACI96481.1 KQ435776 KOX74885.1 FJ219195 FJ219197 ACI96491.1 ACI96493.1 FJ219180 ACI96476.1 FJ219191 ACI96487.1 FJ219208 ACI96504.1 FJ219203 ACI96499.1 FJ219188 FJ219196 FJ219198 FJ219210 ACI96484.1 ACI96492.1 ACI96494.1 ACI96506.1 FJ219192 ACI96488.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000037510
UP000007151
UP000053268
+ More
UP000053240 UP000002320 UP000075882 UP000092553 UP000075840 UP000076407 UP000075881 UP000007062 UP000075903 UP000075902 UP000002282 UP000069940 UP000249989 UP000030765 UP000037069 UP000092443 UP000008744 UP000001819 UP000092460 UP000008820 UP000076502 UP000092445 UP000007801 UP000075884 UP000095300 UP000268350 UP000075886 UP000001292 UP000075885 UP000008711 UP000078200 UP000000803 UP000192221 UP000005203 UP000092444 UP000053825 UP000075883 UP000242457 UP000002358 UP000008792 UP000091820 UP000075920 UP000007798 UP000076408 UP000001070 UP000215335 UP000078541 UP000075809 UP000075900 UP000095301 UP000183832 UP000009192 UP000078492 UP000092462 UP000078540 UP000005205 UP000007755 UP000008237 UP000078542 UP000069272 UP000000673 UP000075880 UP000053097 UP000279307 UP000092461 UP000000311 UP000053105
UP000053240 UP000002320 UP000075882 UP000092553 UP000075840 UP000076407 UP000075881 UP000007062 UP000075903 UP000075902 UP000002282 UP000069940 UP000249989 UP000030765 UP000037069 UP000092443 UP000008744 UP000001819 UP000092460 UP000008820 UP000076502 UP000092445 UP000007801 UP000075884 UP000095300 UP000268350 UP000075886 UP000001292 UP000075885 UP000008711 UP000078200 UP000000803 UP000192221 UP000005203 UP000092444 UP000053825 UP000075883 UP000242457 UP000002358 UP000008792 UP000091820 UP000075920 UP000007798 UP000076408 UP000001070 UP000215335 UP000078541 UP000075809 UP000075900 UP000095301 UP000183832 UP000009192 UP000078492 UP000092462 UP000078540 UP000005205 UP000007755 UP000008237 UP000078542 UP000069272 UP000000673 UP000075880 UP000053097 UP000279307 UP000092461 UP000000311 UP000053105
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9JAK0
A1E130
A0A3S2LKH3
A0A2A4J703
A0A2H1WLZ7
A0A0L7LN46
+ More
A0A212F9G4 A0A194PE52 A0A194QKT3 A0A1Q3FEE7 A0A1Q3FEI2 A0A336LWZ5 B0X1T3 A0A182LQH0 A0A0M4F5S2 A0A182HZG2 A0A182X398 A0A182JT54 Q7PTK6 A0A182VN44 A0A182UGG8 B4PPJ6 A0A182G315 A0A084W2N5 A0A0L0BYK3 A0A1A9YQ61 B4G304 Q297T2 A0A1B0B2Z0 A0A0A1X047 Q16RX2 A0A154PQN6 A0A1A9ZSV7 B3LWX8 A0A1S4FRB8 A0A182NMP0 A0A1I8PCQ8 A0A0C9QWK4 A0A3B0JL46 A0A0K8U4R0 A0A182R079 B4HZF4 A0A182PCR1 A0A3B0K7S6 A0A034W445 B3P5C7 A0A1A9VDP2 A0A0B4LI25 P20480 A0A1W4UC82 W8C7M3 A0A088A154 A0A1B0FL90 A0A0L7R3V3 A0A182LWB3 A0A0L7QW86 A0A2A3ELK3 K7J2W4 B4M0Q0 A0A1A9W5P3 A0A182VQ27 B4NBM4 A0A182Y4Q2 B4JRJ6 A0A232F0M7 A0A195EVA0 A0A151X3M4 A0A182RH46 A0A1I8MBC3 A0A1J1HYB5 B4KAN3 A0A195E5Y2 A0A1B0DP57 A0A151HZ56 A0A158NR31 F4WPB2 E2C5K6 A0A151IN90 A0A182FSW8 W5J5H6 A0A182JKR6 A0A026WWH4 A0A3L8D998 A0A1B0EWA3 A0A1B0BZB5 E2AJ68 A0A1A9XDN0 A0A1B0G3E0 B6UXG3 B6UXD7 A0A0M9A0I4 B6UXE7 B6UXD2 A0A1A9UMT1 B6UXE3 B6UXG0 B6UXF5 B6UXE0 B6UXE4
A0A212F9G4 A0A194PE52 A0A194QKT3 A0A1Q3FEE7 A0A1Q3FEI2 A0A336LWZ5 B0X1T3 A0A182LQH0 A0A0M4F5S2 A0A182HZG2 A0A182X398 A0A182JT54 Q7PTK6 A0A182VN44 A0A182UGG8 B4PPJ6 A0A182G315 A0A084W2N5 A0A0L0BYK3 A0A1A9YQ61 B4G304 Q297T2 A0A1B0B2Z0 A0A0A1X047 Q16RX2 A0A154PQN6 A0A1A9ZSV7 B3LWX8 A0A1S4FRB8 A0A182NMP0 A0A1I8PCQ8 A0A0C9QWK4 A0A3B0JL46 A0A0K8U4R0 A0A182R079 B4HZF4 A0A182PCR1 A0A3B0K7S6 A0A034W445 B3P5C7 A0A1A9VDP2 A0A0B4LI25 P20480 A0A1W4UC82 W8C7M3 A0A088A154 A0A1B0FL90 A0A0L7R3V3 A0A182LWB3 A0A0L7QW86 A0A2A3ELK3 K7J2W4 B4M0Q0 A0A1A9W5P3 A0A182VQ27 B4NBM4 A0A182Y4Q2 B4JRJ6 A0A232F0M7 A0A195EVA0 A0A151X3M4 A0A182RH46 A0A1I8MBC3 A0A1J1HYB5 B4KAN3 A0A195E5Y2 A0A1B0DP57 A0A151HZ56 A0A158NR31 F4WPB2 E2C5K6 A0A151IN90 A0A182FSW8 W5J5H6 A0A182JKR6 A0A026WWH4 A0A3L8D998 A0A1B0EWA3 A0A1B0BZB5 E2AJ68 A0A1A9XDN0 A0A1B0G3E0 B6UXG3 B6UXD7 A0A0M9A0I4 B6UXE7 B6UXD2 A0A1A9UMT1 B6UXE3 B6UXG0 B6UXF5 B6UXE0 B6UXE4
PDB
5W3D
E-value=2.3072e-97,
Score=910
Ontologies
GO
GO:0008017
GO:0005524
GO:0003777
GO:0007018
GO:0003774
GO:0005874
GO:0016887
GO:0090307
GO:0005634
GO:0005871
GO:0072686
GO:0000022
GO:0072687
GO:0007100
GO:0001578
GO:0031534
GO:0051028
GO:0032837
GO:1901673
GO:0090306
GO:0048471
GO:0030981
GO:0005813
GO:1990498
GO:0042803
GO:0032888
GO:0000212
GO:0007056
GO:0008569
GO:0051301
GO:0007052
GO:0005819
GO:0005829
GO:0007059
GO:0007051
GO:0016021
GO:0006171
GO:0035076
GO:0043401
GO:0008270
GO:0003700
GO:0043565
PANTHER
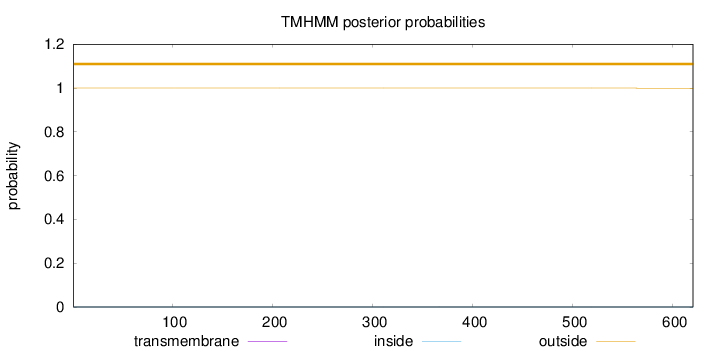
Topology
Subcellular location
Length:
620
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00146
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00034
outside
1 - 620
Population Genetic Test Statistics
Pi
238.292777
Theta
187.642499
Tajima's D
0.743569
CLR
0.165879
CSRT
0.584470776461177
Interpretation
Uncertain
