Gene
KWMTBOMO05645
Pre Gene Modal
BGIBMGA009298
Annotation
PREDICTED:_inositol-trisphosphate_3-kinase_A_[Bombyx_mori]
Full name
Kinase
Location in the cell
Mitochondrial Reliability : 2.94
Sequence
CDS
ATGAACAGTGTTTCCCGTCAATCAATGCTGAGCCGTCTCGGCCCCGGCGGATTACTCTACGCAGCAAATAAAATTTTAGCGGTTGCTGTGCCTTCTAAAGAGGAAGACCGCTGGCATACCAAAGATGGTACCCTATTCAAGCGTTGGCGTCGCACCACGCCGCCCGTGGAACCCGTTCCCACGCCAACTCCTACGGCACCTCCTGCACAGCCTTCGGAACTGTTGAAGTTCTTAGCTATTAACGCTTTGGAACTATCGGCGCCAGCTTCAGATGCGTTACTGAAATCTCGTTCGTCAGAGTGGTTCCAGCTAGCAGGCCATCCTGGATCATTAGCTCCCGCTGGACCCGGTACAGTGTGGAAACGTCGTGCTGCGGGCGACCACCCCGGCCACAACCCTGAACGAGATGCTTACGAAGCCCTTGCAGCTTGTCCTCATATGCGTGGCGTTATTCCTCGCTACTATCGTGAATTGGAATACGACGGTGAACGTTTCATTGAACTTCAAGATCTTCTTCACGGCTTTCGCAATCCTCATGTCATGGACATTAAAATGGGAACCCGGACATTCCTAGAAGATGAAGTCAGCAATGCCCATGCACGTGCCGATCTTTATGAGAAGATGGTCCGTCTAGACCCTAACGCACCCACGGAGTCTGAACATGCCGCCCGAGCTGTCACTAAGCTCCGCTATATGCAGTTTCGTGAACAACGTTCATCTTCTGCCGAGCAAGGTTTTCGTATTGAAGCAGTCAAGTTACCCGGCGAACCTGCCTTGACCGACCTACAGAAAATTCGCGAACCCCACCAATTGGCCGCTACAGTGGCACGTTTCCTTGGGAATGATGAACGAGCACGAAGGGAGATTGCCTCGCGTCTTAGAGAGATCAGAGATCATTTTGAACGTTCGGATTTCTTTAAAAGCCACGAAATTGTTGGCAGCAGCATCTTCATCATTTACGATGATGAACGAGTTGGAGCTTGGCTCATAGATTTCGCAAAAACAAGGAAAATCCCGACAGGCTCGCAAGTCAATCATCGTGCACCGTGGCAACAAGGCAATCATGAGGAAGGCTTCTTGTTTGGGTTAGATAGATTGATACAAACTATAGAAACGGCAAAACTGTCTCCTCAAGGCGATGAAGTTGTGGAAGAACCAGATGTATCACGTTGA
Protein
MNSVSRQSMLSRLGPGGLLYAANKILAVAVPSKEEDRWHTKDGTLFKRWRRTTPPVEPVPTPTPTAPPAQPSELLKFLAINALELSAPASDALLKSRSSEWFQLAGHPGSLAPAGPGTVWKRRAAGDHPGHNPERDAYEALAACPHMRGVIPRYYRELEYDGERFIELQDLLHGFRNPHVMDIKMGTRTFLEDEVSNAHARADLYEKMVRLDPNAPTESEHAARAVTKLRYMQFREQRSSSAEQGFRIEAVKLPGEPALTDLQKIREPHQLAATVARFLGNDERARREIASRLREIRDHFERSDFFKSHEIVGSSIFIIYDDERVGAWLIDFAKTRKIPTGSQVNHRAPWQQGNHEEGFLFGLDRLIQTIETAKLSPQGDEVVEEPDVSR
Summary
Similarity
Belongs to the inositol phosphokinase (IPK) family.
Feature
chain Kinase
Uniprot
A0A2H1X2D1
A0A2A4J7Y0
H9JIE8
A0A3S2PE09
A0A194QKP8
A0A194PEU4
+ More
A0A212F9G6 I4DL82 A0A0L7LNG3 A0A2A4J6W7 S4PHA2 I4DRX2 A0A1E1WL05 A0A1W4WF98 A0A1Y1M1W0 A0A310SJ32 A0A0J7KGH6 A0A1W4WG97 A0A087ZR17 D6WZL6 A0A026WS91 A0A0L7QLN5 A0A2J7QXP7 A0A2A3E7N6 A0A2J7QXM7 A0A0N0BK23 A0A195FN95 E2B9K1 A0A158NF98 A0A195E884 A0A195AVQ2 A0A151WHW4 A0A232EPD0 E9IWI7 F4WEL9 E2AE25 A0A195C460 A0A0T6BGP9 A0A0K8SDB4 A0A0A9Z6A4 A0A0K8SCP7 A0A0A9Z4L4 A0A154PHH2 A0A0C9R802 A0A1B6D9K3 A0A1B6C0Z7 A0A0C9PP17 A0A0C9R4M8 A0A0C9RGC2 A0A0P4VQ78 A0A023F4E9 E0VIU1 A0A069DSW2 A0A1B6FJX8 A0A0V0G6R9 A0A224XEF4 A0A1B6GLG9 A0A2S2P3K2 A0A2R7VWP8 A0A2S2R1T3 A0A2H8TPL3 J9JYH7 A0A0P5IAF8 A0A0P5ALX0 A0A0P5BAF5 A0A0P5JLD8 A0A0P5RJE7 A0A0P4XPC4 A0A162P2B7 A0A0P5NB11 A0A0N8A339 A0A0P4YT11 A0A0P4Z685 A0A0P5Y2K6 A0A0P5D9T6 A0A0P6GYR8 A0A0P5XCC4 A0A0P6DYA8 T1JEJ5 A0A0P4W5Z2 A0A1I8N270 A0A1B0GIS4 A0A210QRJ2 A0A1I8N294 A0A1L8DYS6 A0A1B0GMU6 A0A1I8PHU8 W8AZM6 A0A0R3NRR6 A0A182JKZ2 A0A2M3Z5G5 A0A2M3Z579 A0A087UZG8 A0A2M3ZH65 A0A2P2I5M4 A0A023EQE3 A0A0R3NYB9 A0A2M4AAS0 T1E9Y2
A0A212F9G6 I4DL82 A0A0L7LNG3 A0A2A4J6W7 S4PHA2 I4DRX2 A0A1E1WL05 A0A1W4WF98 A0A1Y1M1W0 A0A310SJ32 A0A0J7KGH6 A0A1W4WG97 A0A087ZR17 D6WZL6 A0A026WS91 A0A0L7QLN5 A0A2J7QXP7 A0A2A3E7N6 A0A2J7QXM7 A0A0N0BK23 A0A195FN95 E2B9K1 A0A158NF98 A0A195E884 A0A195AVQ2 A0A151WHW4 A0A232EPD0 E9IWI7 F4WEL9 E2AE25 A0A195C460 A0A0T6BGP9 A0A0K8SDB4 A0A0A9Z6A4 A0A0K8SCP7 A0A0A9Z4L4 A0A154PHH2 A0A0C9R802 A0A1B6D9K3 A0A1B6C0Z7 A0A0C9PP17 A0A0C9R4M8 A0A0C9RGC2 A0A0P4VQ78 A0A023F4E9 E0VIU1 A0A069DSW2 A0A1B6FJX8 A0A0V0G6R9 A0A224XEF4 A0A1B6GLG9 A0A2S2P3K2 A0A2R7VWP8 A0A2S2R1T3 A0A2H8TPL3 J9JYH7 A0A0P5IAF8 A0A0P5ALX0 A0A0P5BAF5 A0A0P5JLD8 A0A0P5RJE7 A0A0P4XPC4 A0A162P2B7 A0A0P5NB11 A0A0N8A339 A0A0P4YT11 A0A0P4Z685 A0A0P5Y2K6 A0A0P5D9T6 A0A0P6GYR8 A0A0P5XCC4 A0A0P6DYA8 T1JEJ5 A0A0P4W5Z2 A0A1I8N270 A0A1B0GIS4 A0A210QRJ2 A0A1I8N294 A0A1L8DYS6 A0A1B0GMU6 A0A1I8PHU8 W8AZM6 A0A0R3NRR6 A0A182JKZ2 A0A2M3Z5G5 A0A2M3Z579 A0A087UZG8 A0A2M3ZH65 A0A2P2I5M4 A0A023EQE3 A0A0R3NYB9 A0A2M4AAS0 T1E9Y2
EC Number
2.7.-.-
Pubmed
EMBL
ODYU01012877
SOQ59398.1
NWSH01002607
PCG67866.1
BABH01022489
BABH01022490
+ More
RSAL01000080 RVE48612.1 KQ461198 KPJ06138.1 KQ459606 KPI91543.1 AGBW02009576 OWR50396.1 AK402050 BAM18672.1 JTDY01000483 KOB76975.1 PCG67867.1 GAIX01003347 JAA89213.1 AK405310 BAM20662.1 GDQN01003399 JAT87655.1 GEZM01042506 GEZM01042505 JAV79814.1 KQ759804 OAD62798.1 LBMM01007867 KMQ89349.1 KQ971372 EFA10445.2 KK107119 QOIP01000009 EZA58531.1 RLU18686.1 KQ414924 KOC59426.1 NEVH01009378 PNF33334.1 KZ288344 PBC27718.1 PNF33335.1 KQ435709 KOX79988.1 KQ981490 KYN41359.1 GL446556 EFN87634.1 ADTU01013710 ADTU01013711 KQ979479 KYN21331.1 KQ976736 KYM76049.1 KQ983106 KYQ47410.1 NNAY01002948 OXU20239.1 GL766526 EFZ15076.1 GL888103 EGI67382.1 GL438827 EFN68258.1 KQ978287 KYM95639.1 LJIG01000507 KRT86433.1 GBRD01014757 JAG51069.1 GBHO01004193 GBHO01004191 JAG39411.1 JAG39413.1 GBRD01014759 JAG51067.1 GBHO01004192 GBHO01004190 JAG39412.1 JAG39414.1 KQ434907 KZC11299.1 GBYB01002906 JAG72673.1 GEDC01014988 JAS22310.1 GEDC01030122 JAS07176.1 GBYB01002908 JAG72675.1 GBYB01002905 JAG72672.1 GBYB01007400 JAG77167.1 GDKW01002024 JAI54571.1 GBBI01002818 JAC15894.1 DS235206 EEB13297.1 GBGD01001854 JAC87035.1 GECZ01019277 JAS50492.1 GECL01002485 JAP03639.1 GFTR01005581 JAW10845.1 GECZ01006483 JAS63286.1 GGMR01011428 MBY24047.1 KK854092 PTY11110.1 GGMS01014109 MBY83312.1 GFXV01004125 MBW15930.1 ABLF02027485 GDIQ01223000 JAK28725.1 GDIP01197429 JAJ25973.1 GDIP01187224 JAJ36178.1 GDIQ01223001 JAK28724.1 GDIP01175236 GDIQ01206502 GDIQ01186850 GDIQ01146786 GDIQ01101693 JAJ48166.1 JAL50033.1 GDIP01239686 JAI83715.1 LRGB01000512 KZS18459.1 GDIQ01148192 JAL03534.1 GDIP01187225 JAJ36177.1 GDIP01222974 JAJ00428.1 GDIP01219822 JAJ03580.1 GDIP01063850 JAM39865.1 GDIP01163332 JAJ60070.1 GDIQ01026759 JAN67978.1 GDIP01074251 JAM29464.1 GDIQ01070462 JAN24275.1 JH432122 GDRN01087491 JAI61028.1 AJWK01015364 AJWK01015365 AJWK01015366 NEDP02002292 OWF51308.1 GFDF01002461 JAV11623.1 AJVK01003763 GAMC01016237 JAB90318.1 CH379060 KRT03810.1 GGFM01002937 MBW23688.1 GGFM01002928 MBW23679.1 KK122468 KFM82757.1 GGFM01007123 MBW27874.1 IACF01003700 LAB69311.1 GAPW01002303 JAC11295.1 KRT03811.1 GGFK01004562 MBW37883.1 GAMD01001508 JAB00083.1
RSAL01000080 RVE48612.1 KQ461198 KPJ06138.1 KQ459606 KPI91543.1 AGBW02009576 OWR50396.1 AK402050 BAM18672.1 JTDY01000483 KOB76975.1 PCG67867.1 GAIX01003347 JAA89213.1 AK405310 BAM20662.1 GDQN01003399 JAT87655.1 GEZM01042506 GEZM01042505 JAV79814.1 KQ759804 OAD62798.1 LBMM01007867 KMQ89349.1 KQ971372 EFA10445.2 KK107119 QOIP01000009 EZA58531.1 RLU18686.1 KQ414924 KOC59426.1 NEVH01009378 PNF33334.1 KZ288344 PBC27718.1 PNF33335.1 KQ435709 KOX79988.1 KQ981490 KYN41359.1 GL446556 EFN87634.1 ADTU01013710 ADTU01013711 KQ979479 KYN21331.1 KQ976736 KYM76049.1 KQ983106 KYQ47410.1 NNAY01002948 OXU20239.1 GL766526 EFZ15076.1 GL888103 EGI67382.1 GL438827 EFN68258.1 KQ978287 KYM95639.1 LJIG01000507 KRT86433.1 GBRD01014757 JAG51069.1 GBHO01004193 GBHO01004191 JAG39411.1 JAG39413.1 GBRD01014759 JAG51067.1 GBHO01004192 GBHO01004190 JAG39412.1 JAG39414.1 KQ434907 KZC11299.1 GBYB01002906 JAG72673.1 GEDC01014988 JAS22310.1 GEDC01030122 JAS07176.1 GBYB01002908 JAG72675.1 GBYB01002905 JAG72672.1 GBYB01007400 JAG77167.1 GDKW01002024 JAI54571.1 GBBI01002818 JAC15894.1 DS235206 EEB13297.1 GBGD01001854 JAC87035.1 GECZ01019277 JAS50492.1 GECL01002485 JAP03639.1 GFTR01005581 JAW10845.1 GECZ01006483 JAS63286.1 GGMR01011428 MBY24047.1 KK854092 PTY11110.1 GGMS01014109 MBY83312.1 GFXV01004125 MBW15930.1 ABLF02027485 GDIQ01223000 JAK28725.1 GDIP01197429 JAJ25973.1 GDIP01187224 JAJ36178.1 GDIQ01223001 JAK28724.1 GDIP01175236 GDIQ01206502 GDIQ01186850 GDIQ01146786 GDIQ01101693 JAJ48166.1 JAL50033.1 GDIP01239686 JAI83715.1 LRGB01000512 KZS18459.1 GDIQ01148192 JAL03534.1 GDIP01187225 JAJ36177.1 GDIP01222974 JAJ00428.1 GDIP01219822 JAJ03580.1 GDIP01063850 JAM39865.1 GDIP01163332 JAJ60070.1 GDIQ01026759 JAN67978.1 GDIP01074251 JAM29464.1 GDIQ01070462 JAN24275.1 JH432122 GDRN01087491 JAI61028.1 AJWK01015364 AJWK01015365 AJWK01015366 NEDP02002292 OWF51308.1 GFDF01002461 JAV11623.1 AJVK01003763 GAMC01016237 JAB90318.1 CH379060 KRT03810.1 GGFM01002937 MBW23688.1 GGFM01002928 MBW23679.1 KK122468 KFM82757.1 GGFM01007123 MBW27874.1 IACF01003700 LAB69311.1 GAPW01002303 JAC11295.1 KRT03811.1 GGFK01004562 MBW37883.1 GAMD01001508 JAB00083.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000192223 UP000036403 UP000005203 UP000007266 UP000053097 UP000279307 UP000053825 UP000235965 UP000242457 UP000053105 UP000078541 UP000008237 UP000005205 UP000078492 UP000078540 UP000075809 UP000215335 UP000007755 UP000000311 UP000078542 UP000076502 UP000009046 UP000007819 UP000076858 UP000095301 UP000092461 UP000242188 UP000092462 UP000095300 UP000001819 UP000075880 UP000054359
UP000037510 UP000192223 UP000036403 UP000005203 UP000007266 UP000053097 UP000279307 UP000053825 UP000235965 UP000242457 UP000053105 UP000078541 UP000008237 UP000005205 UP000078492 UP000078540 UP000075809 UP000215335 UP000007755 UP000000311 UP000078542 UP000076502 UP000009046 UP000007819 UP000076858 UP000095301 UP000092461 UP000242188 UP000092462 UP000095300 UP000001819 UP000075880 UP000054359
PRIDE
Gene 3D
ProteinModelPortal
A0A2H1X2D1
A0A2A4J7Y0
H9JIE8
A0A3S2PE09
A0A194QKP8
A0A194PEU4
+ More
A0A212F9G6 I4DL82 A0A0L7LNG3 A0A2A4J6W7 S4PHA2 I4DRX2 A0A1E1WL05 A0A1W4WF98 A0A1Y1M1W0 A0A310SJ32 A0A0J7KGH6 A0A1W4WG97 A0A087ZR17 D6WZL6 A0A026WS91 A0A0L7QLN5 A0A2J7QXP7 A0A2A3E7N6 A0A2J7QXM7 A0A0N0BK23 A0A195FN95 E2B9K1 A0A158NF98 A0A195E884 A0A195AVQ2 A0A151WHW4 A0A232EPD0 E9IWI7 F4WEL9 E2AE25 A0A195C460 A0A0T6BGP9 A0A0K8SDB4 A0A0A9Z6A4 A0A0K8SCP7 A0A0A9Z4L4 A0A154PHH2 A0A0C9R802 A0A1B6D9K3 A0A1B6C0Z7 A0A0C9PP17 A0A0C9R4M8 A0A0C9RGC2 A0A0P4VQ78 A0A023F4E9 E0VIU1 A0A069DSW2 A0A1B6FJX8 A0A0V0G6R9 A0A224XEF4 A0A1B6GLG9 A0A2S2P3K2 A0A2R7VWP8 A0A2S2R1T3 A0A2H8TPL3 J9JYH7 A0A0P5IAF8 A0A0P5ALX0 A0A0P5BAF5 A0A0P5JLD8 A0A0P5RJE7 A0A0P4XPC4 A0A162P2B7 A0A0P5NB11 A0A0N8A339 A0A0P4YT11 A0A0P4Z685 A0A0P5Y2K6 A0A0P5D9T6 A0A0P6GYR8 A0A0P5XCC4 A0A0P6DYA8 T1JEJ5 A0A0P4W5Z2 A0A1I8N270 A0A1B0GIS4 A0A210QRJ2 A0A1I8N294 A0A1L8DYS6 A0A1B0GMU6 A0A1I8PHU8 W8AZM6 A0A0R3NRR6 A0A182JKZ2 A0A2M3Z5G5 A0A2M3Z579 A0A087UZG8 A0A2M3ZH65 A0A2P2I5M4 A0A023EQE3 A0A0R3NYB9 A0A2M4AAS0 T1E9Y2
A0A212F9G6 I4DL82 A0A0L7LNG3 A0A2A4J6W7 S4PHA2 I4DRX2 A0A1E1WL05 A0A1W4WF98 A0A1Y1M1W0 A0A310SJ32 A0A0J7KGH6 A0A1W4WG97 A0A087ZR17 D6WZL6 A0A026WS91 A0A0L7QLN5 A0A2J7QXP7 A0A2A3E7N6 A0A2J7QXM7 A0A0N0BK23 A0A195FN95 E2B9K1 A0A158NF98 A0A195E884 A0A195AVQ2 A0A151WHW4 A0A232EPD0 E9IWI7 F4WEL9 E2AE25 A0A195C460 A0A0T6BGP9 A0A0K8SDB4 A0A0A9Z6A4 A0A0K8SCP7 A0A0A9Z4L4 A0A154PHH2 A0A0C9R802 A0A1B6D9K3 A0A1B6C0Z7 A0A0C9PP17 A0A0C9R4M8 A0A0C9RGC2 A0A0P4VQ78 A0A023F4E9 E0VIU1 A0A069DSW2 A0A1B6FJX8 A0A0V0G6R9 A0A224XEF4 A0A1B6GLG9 A0A2S2P3K2 A0A2R7VWP8 A0A2S2R1T3 A0A2H8TPL3 J9JYH7 A0A0P5IAF8 A0A0P5ALX0 A0A0P5BAF5 A0A0P5JLD8 A0A0P5RJE7 A0A0P4XPC4 A0A162P2B7 A0A0P5NB11 A0A0N8A339 A0A0P4YT11 A0A0P4Z685 A0A0P5Y2K6 A0A0P5D9T6 A0A0P6GYR8 A0A0P5XCC4 A0A0P6DYA8 T1JEJ5 A0A0P4W5Z2 A0A1I8N270 A0A1B0GIS4 A0A210QRJ2 A0A1I8N294 A0A1L8DYS6 A0A1B0GMU6 A0A1I8PHU8 W8AZM6 A0A0R3NRR6 A0A182JKZ2 A0A2M3Z5G5 A0A2M3Z579 A0A087UZG8 A0A2M3ZH65 A0A2P2I5M4 A0A023EQE3 A0A0R3NYB9 A0A2M4AAS0 T1E9Y2
PDB
1W2F
E-value=4.56462e-61,
Score=594
Ontologies
PATHWAY
GO
PANTHER
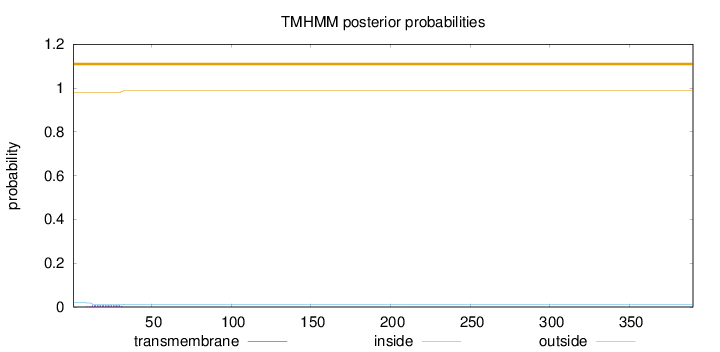
Topology
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16928
Exp number, first 60 AAs:
0.1686
Total prob of N-in:
0.02005
outside
1 - 390
Population Genetic Test Statistics
Pi
289.607738
Theta
234.27796
Tajima's D
0.892104
CLR
119.142471
CSRT
0.632768361581921
Interpretation
Uncertain
