Gene
KWMTBOMO05637
Pre Gene Modal
BGIBMGA006784
Annotation
PREDICTED:_rotatin-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.026
Sequence
CDS
ATGAGTAAACCTGAGATTATAATTAATTACATTCAGAAGCTTAGACATCCAATAAAAGAGATTCGTGAACGCTCTCTTCAATCATTAAAATCCAAATTCAAGTTGGGATGGGAGTTCGATGATGAGATTTCGGGAACCAGGTTTCTATTGGAATCCTTAATGGAATGGTTCAATGACGAGAAACCGCTTCTACAGCGAGAAGCTTTAGATTTGCTACACTCAGTAATAAAGACAAAATCTGGGACATACATCGTACAAGAAACAGGCTTAAACAATTTGGTTACAAAAATACAGAAATTTAAAGATAAAATAGATAAGGATGCATTAAATATCTATGAAGACATTATGGAGAGTCTGACCTTCTTAAACACAGTGTCTTCAGAAGAAAATTTGAATATTCCAAGACTAAGTTTATCTGAAGAAATGAGAGCTGAAAGTTGTGGGAGATCAAGCAGCAGTTACTATAATTTAGCTTCAAGCACACAGAGTTCAAAAGAATCATCGGTTTTAAACATATGTTTAGATGTCAACGGGCAAAGTTCGAATTCTGATGATGGTGTAAAAGTGCTGCTATTTCCATGGGCTGATCTAAGTCAATCAGATATTAGACCCATTCTTTTAATCGAGGATGCTCTAAGGCTACTTCGTTCTACCAGACGATGTTGCAGATTTATCCGAGATGTATTTTTGCGCGATTTTCCTGCAGAAATATTCTTAAATCGTCCTCAAATTATAAAGAATCTTTGGTCAATGATTGATGGAAATAATAAAACTCATTCTGAAGAGGCACTACATGTACTATTTCACATAACGTCTGCATTGCGGTCCAGGCTCTTACAATTAAAGTCTTTTGACCTCAAATACAGCATCCAGTTTACAGGCGATGATAAAATCTCGCAGAATGATGTTAATTTAGAGCTGGAACAAATTACAGGAGATGACTTTTCTGGGTCGTTAGAGGAAGATAAACTCATAGCTTTAAAGCAATTACCTGCTCCTATTTTTACTTTAGAATCCCTTCATAGTATATTTTCTACAATGAATAGGAATTTAACTTTTTTTGAACAGAATGAATTAAGTGACATTCTTGCCTGGAAAGAACTAAATACATTCTTGCAATTAATCGAGGTCCTTGTTGGATTGCTTTTCGACTGTGTTACGGAAAAATTCTGGTCTGTTGACCATACTTCGAAGACTCACAGAGATATCTCTCACAAAAGTTGTATGGTCATGCGCGTATTAGGGGACATACTTACGAAGTACAGAGAATCATTTTTCAATGACACAGCAGCGGAGCGTTATCGAGTTGCTTGGTTACGTTTGATATTTTGTGGGGAAAGATTTCTTAATTGGGCGAGAAACTCAGCTTTACCACCCTCATCTCTGGTATCCGCCGTACAAGCTGCGCAACTAGATCCAGCGGTGGACTTATTTTACCCTGAATTGAGTAATAGAATATCAATTGTTTTGCAGAATGTGAATCTGTCTGTAGACCAAGAATACAAAAGTAAATACCGAGAGTTGAAAATGCTATTCTCCAGCATGGAGCACGCTGTAAACTTTATGAAGAATAAGAATTCGTGTCGTAATACAACCAAAGTACTGACCGTCATAACAGATGCGCTTCCTACTTTGGAATTGATTCAAAGCGAGACTTTTCTAAATGACGTTATAAATGTGTTATTGATTAAAACTAAAGACTTTGTTATGGATGAAAACGATTGGTCCAAGGCGAGGAATATAGCTTTGAAATTGATGGCACACAATTCGGAGTGGGTTCGTGCCAAATTCTATGAGAGAATGGCGGATATGGTGAAATCAATTTTAGTCGATGAAACCAACCAATCGGTTAATGAAAAAAGTTTGACTTTGATATGTGATGTGGGAGTTTTGACTGAGATATGCTGTCACGGTCTTTCTTCTAAATCAAAGGAGGTAGAGGAACATGCTACGGATGTTATGCTGTATCTGCTGCGAGGGCGGCTGGTGCTCAGTGACGGTTGCTGGTGGCGTCTGATGGCCAGCTTGTTGCCCGTCCTACCGCTGCTACACGTATACGCTGCACACGAAACACATCTTGGACGAGCGATTTGCAAGTCACTGGAACGCGACATCGCCAATTGCATGGGCGTGTCGAACGCGGACTTGATATCGGGCTTGGTGCGGCTTTTATTCGTGCGGTGCGCTACGGTACAGTTAGACGCCGCCCACTCCTTATGTCGTTTACTAGACGACGAAAGATACTTACCTCCGCAGCAATCTCTAAGAGTTGATGTTTTACTAAGCGCATTAAGAAGAGTGAAAACTCAAGAGTTCAATTTGGATACTTCTAGTTCACCCACAAAGATGATACAGACGACCGCCCTATTACAAATGTTGGACGTTTTAAAACAAGGCATCGTGCTGGACGAGCGCGGCGCCGAGCTCGTGGCGCGCCCCCCGCTGCGGCCCGCGCTCGAGCCGTCGCTGCGCCGCACCACCCTGCAGCAGCTCGCGCTCGTCATGCGGCAACAACAACTGCACGAAACATTCATGGAGTACGACGGCCTTCGAGTTGTCGTGGCTACGTTGAGGATGTCCTTACTGGTGGATGACTATCTTGCGTTTCCGGAGTGCGCGGTCAGCTGCGTGTCGGTGCTAAACAGTGTGTGCTTCACGGCTCGTCACGCCTTGGCCAAGATACCTGACCTATTTTTGCTATTGTTGAGAGTGATATTGGTGTTTCCAACAAATGAAGCTGCAGTGGCTATGACCGCACAAGTGCTGGCTCTAGTGGCGTGGGCCGGATTTGTATTACAAGAGCTAGATTCTGCTCGACGTCGCGTACCAGCTCTGCCATTAAACGTGACGGACCGGATATCTCTGCCTTTTAACGTCCACAGCTATTGGAGCACTAGCCCTAATGCTGAACATTCATACATTGAGTGGTTGTTAGGGGAGGAACAGTGGCGTGCATGTATCCGCGTGCGCTGGTGGTGCACGTACGCGGGGCGCGCGTGCGTGCTGCAGGGGCCGAGCGCCGCCGCCGCGTGCCCCGCCCCGCTGGGCCTGCAGCCCGCCGCGCGCGACGTGGCCGCGCTGAGGACGGCGTGCCCGCAGTACGCGTGCGGCGCCGCCCTGCTCGCGCTCGAGAACGCGACCTCGCATGCCCAGTGA
Protein
MSKPEIIINYIQKLRHPIKEIRERSLQSLKSKFKLGWEFDDEISGTRFLLESLMEWFNDEKPLLQREALDLLHSVIKTKSGTYIVQETGLNNLVTKIQKFKDKIDKDALNIYEDIMESLTFLNTVSSEENLNIPRLSLSEEMRAESCGRSSSSYYNLASSTQSSKESSVLNICLDVNGQSSNSDDGVKVLLFPWADLSQSDIRPILLIEDALRLLRSTRRCCRFIRDVFLRDFPAEIFLNRPQIIKNLWSMIDGNNKTHSEEALHVLFHITSALRSRLLQLKSFDLKYSIQFTGDDKISQNDVNLELEQITGDDFSGSLEEDKLIALKQLPAPIFTLESLHSIFSTMNRNLTFFEQNELSDILAWKELNTFLQLIEVLVGLLFDCVTEKFWSVDHTSKTHRDISHKSCMVMRVLGDILTKYRESFFNDTAAERYRVAWLRLIFCGERFLNWARNSALPPSSLVSAVQAAQLDPAVDLFYPELSNRISIVLQNVNLSVDQEYKSKYRELKMLFSSMEHAVNFMKNKNSCRNTTKVLTVITDALPTLELIQSETFLNDVINVLLIKTKDFVMDENDWSKARNIALKLMAHNSEWVRAKFYERMADMVKSILVDETNQSVNEKSLTLICDVGVLTEICCHGLSSKSKEVEEHATDVMLYLLRGRLVLSDGCWWRLMASLLPVLPLLHVYAAHETHLGRAICKSLERDIANCMGVSNADLISGLVRLLFVRCATVQLDAAHSLCRLLDDERYLPPQQSLRVDVLLSALRRVKTQEFNLDTSSSPTKMIQTTALLQMLDVLKQGIVLDERGAELVARPPLRPALEPSLRRTTLQQLALVMRQQQLHETFMEYDGLRVVVATLRMSLLVDDYLAFPECAVSCVSVLNSVCFTARHALAKIPDLFLLLLRVILVFPTNEAAVAMTAQVLALVAWAGFVLQELDSARRRVPALPLNVTDRISLPFNVHSYWSTSPNAEHSYIEWLLGEEQWRACIRVRWWCTYAGRACVLQGPSAAAACPAPLGLQPAARDVAALRTACPQYACGAALLALENATSHAQ
Summary
Uniprot
H9JB89
A0A2A4J928
A0A212EUN4
A0A194QMF9
A0A194PE57
A0A139WBK6
+ More
A0A195EP54 A0A158NEG1 A0A195BV69 A0A154PRC5 A0A195FKC7 A0A151XJ89 F4WVJ4 A0A310SXI6 E9ID30 A0A0L7QLD8 E2B9K2 K7IRQ8 A0A232FD48 E0VIU0 A0A087ZR13 A0A0C9RCN8 A0A2A3E951 A0A0M9A9J8 A0A1Y1NBB4 A0A195CQA9 V4AU31 W5M728 A0A3L8E4K6 I3KA67 F7CVJ3 G3WGH6 A0A2U9CXV1 A0A3Q1GHK3 A0A1W5BJ36 A0A0L8G8T4 G3WGH8 C4TFE0 A0A3P9D2Y4 C4TFD9 A0A3Q1B8S3 H2LUM2 C4TFD6 C4TFE2 C4TFE1 E6ZIL3 C4TFD8 C4TFD7 G3WGH7 A0A3Q4G585 A0A3Q2X1X5 A0A3B5B6K1 A0A3Q3FFD4
A0A195EP54 A0A158NEG1 A0A195BV69 A0A154PRC5 A0A195FKC7 A0A151XJ89 F4WVJ4 A0A310SXI6 E9ID30 A0A0L7QLD8 E2B9K2 K7IRQ8 A0A232FD48 E0VIU0 A0A087ZR13 A0A0C9RCN8 A0A2A3E951 A0A0M9A9J8 A0A1Y1NBB4 A0A195CQA9 V4AU31 W5M728 A0A3L8E4K6 I3KA67 F7CVJ3 G3WGH6 A0A2U9CXV1 A0A3Q1GHK3 A0A1W5BJ36 A0A0L8G8T4 G3WGH8 C4TFE0 A0A3P9D2Y4 C4TFD9 A0A3Q1B8S3 H2LUM2 C4TFD6 C4TFE2 C4TFE1 E6ZIL3 C4TFD8 C4TFD7 G3WGH7 A0A3Q4G585 A0A3Q2X1X5 A0A3B5B6K1 A0A3Q3FFD4
Pubmed
EMBL
BABH01008785
NWSH01002591
PCG67903.1
AGBW02012332
OWR45199.1
KQ461198
+ More
KPJ06140.1 KQ459606 KPI91547.1 KQ971372 KYB25348.1 KQ978625 KYN29664.1 ADTU01013285 KQ976401 KYM92477.1 KQ435057 KZC14297.1 KQ981490 KYN41120.1 KQ982080 KYQ60397.1 GL888387 EGI61815.1 KQ759804 OAD62796.1 GL762454 EFZ21315.1 KQ414924 KOC59425.1 GL446556 EFN87635.1 NNAY01000408 OXU28605.1 DS235206 EEB13296.1 GBYB01014329 JAG84096.1 KZ288344 PBC27719.1 KQ435709 KOX79990.1 GEZM01007669 JAV95009.1 KQ977444 KYN02662.1 KB201262 ESO98430.1 AHAT01004312 AHAT01004313 AHAT01004314 AHAT01004315 AHAT01004316 AHAT01004317 AHAT01004318 QOIP01000001 RLU27556.1 AERX01010073 AERX01010074 AEFK01062067 AEFK01062068 AEFK01062069 AEFK01062070 AEFK01062071 AEFK01062072 AEFK01062073 AEFK01062074 AEFK01062075 AEFK01062076 CP026263 AWP20729.1 KQ423217 KOF73283.1 AB435849 BAH66370.1 AB435848 BAH66369.1 AB435845 BAH66366.1 AB435851 BAH66372.1 AB435850 BAH66371.1 FQ310508 CBN81897.1 AB435847 BAH66368.1 AB435846 BAH66367.1
KPJ06140.1 KQ459606 KPI91547.1 KQ971372 KYB25348.1 KQ978625 KYN29664.1 ADTU01013285 KQ976401 KYM92477.1 KQ435057 KZC14297.1 KQ981490 KYN41120.1 KQ982080 KYQ60397.1 GL888387 EGI61815.1 KQ759804 OAD62796.1 GL762454 EFZ21315.1 KQ414924 KOC59425.1 GL446556 EFN87635.1 NNAY01000408 OXU28605.1 DS235206 EEB13296.1 GBYB01014329 JAG84096.1 KZ288344 PBC27719.1 KQ435709 KOX79990.1 GEZM01007669 JAV95009.1 KQ977444 KYN02662.1 KB201262 ESO98430.1 AHAT01004312 AHAT01004313 AHAT01004314 AHAT01004315 AHAT01004316 AHAT01004317 AHAT01004318 QOIP01000001 RLU27556.1 AERX01010073 AERX01010074 AEFK01062067 AEFK01062068 AEFK01062069 AEFK01062070 AEFK01062071 AEFK01062072 AEFK01062073 AEFK01062074 AEFK01062075 AEFK01062076 CP026263 AWP20729.1 KQ423217 KOF73283.1 AB435849 BAH66370.1 AB435848 BAH66369.1 AB435845 BAH66366.1 AB435851 BAH66372.1 AB435850 BAH66371.1 FQ310508 CBN81897.1 AB435847 BAH66368.1 AB435846 BAH66367.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000078492 UP000005205 UP000078540 UP000076502 UP000078541 UP000075809 UP000007755 UP000053825 UP000008237 UP000002358 UP000215335 UP000009046 UP000005203 UP000242457 UP000053105 UP000078542 UP000030746 UP000018468 UP000279307 UP000005207 UP000002280 UP000007648 UP000246464 UP000257200 UP000053454 UP000265160 UP000257160 UP000001038 UP000261580 UP000264840 UP000261400 UP000261660
UP000078492 UP000005205 UP000078540 UP000076502 UP000078541 UP000075809 UP000007755 UP000053825 UP000008237 UP000002358 UP000215335 UP000009046 UP000005203 UP000242457 UP000053105 UP000078542 UP000030746 UP000018468 UP000279307 UP000005207 UP000002280 UP000007648 UP000246464 UP000257200 UP000053454 UP000265160 UP000257160 UP000001038 UP000261580 UP000264840 UP000261400 UP000261660
PRIDE
Interpro
IPR030791
Rotatin
+ More
IPR001506 Peptidase_M12A
IPR029249 Rotatin_N
IPR024079 MetalloPept_cat_dom_sf
IPR016024 ARM-type_fold
IPR002401 Cyt_P450_E_grp-I
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR036396 Cyt_P450_sf
IPR004117 7tm6_olfct_rcpt
IPR011989 ARM-like
IPR018188 RNase_T2_His_AS_1
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013106 Ig_V-set
IPR001506 Peptidase_M12A
IPR029249 Rotatin_N
IPR024079 MetalloPept_cat_dom_sf
IPR016024 ARM-type_fold
IPR002401 Cyt_P450_E_grp-I
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR036396 Cyt_P450_sf
IPR004117 7tm6_olfct_rcpt
IPR011989 ARM-like
IPR018188 RNase_T2_His_AS_1
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013106 Ig_V-set
Gene 3D
ProteinModelPortal
H9JB89
A0A2A4J928
A0A212EUN4
A0A194QMF9
A0A194PE57
A0A139WBK6
+ More
A0A195EP54 A0A158NEG1 A0A195BV69 A0A154PRC5 A0A195FKC7 A0A151XJ89 F4WVJ4 A0A310SXI6 E9ID30 A0A0L7QLD8 E2B9K2 K7IRQ8 A0A232FD48 E0VIU0 A0A087ZR13 A0A0C9RCN8 A0A2A3E951 A0A0M9A9J8 A0A1Y1NBB4 A0A195CQA9 V4AU31 W5M728 A0A3L8E4K6 I3KA67 F7CVJ3 G3WGH6 A0A2U9CXV1 A0A3Q1GHK3 A0A1W5BJ36 A0A0L8G8T4 G3WGH8 C4TFE0 A0A3P9D2Y4 C4TFD9 A0A3Q1B8S3 H2LUM2 C4TFD6 C4TFE2 C4TFE1 E6ZIL3 C4TFD8 C4TFD7 G3WGH7 A0A3Q4G585 A0A3Q2X1X5 A0A3B5B6K1 A0A3Q3FFD4
A0A195EP54 A0A158NEG1 A0A195BV69 A0A154PRC5 A0A195FKC7 A0A151XJ89 F4WVJ4 A0A310SXI6 E9ID30 A0A0L7QLD8 E2B9K2 K7IRQ8 A0A232FD48 E0VIU0 A0A087ZR13 A0A0C9RCN8 A0A2A3E951 A0A0M9A9J8 A0A1Y1NBB4 A0A195CQA9 V4AU31 W5M728 A0A3L8E4K6 I3KA67 F7CVJ3 G3WGH6 A0A2U9CXV1 A0A3Q1GHK3 A0A1W5BJ36 A0A0L8G8T4 G3WGH8 C4TFE0 A0A3P9D2Y4 C4TFD9 A0A3Q1B8S3 H2LUM2 C4TFD6 C4TFE2 C4TFE1 E6ZIL3 C4TFD8 C4TFD7 G3WGH7 A0A3Q4G585 A0A3Q2X1X5 A0A3B5B6K1 A0A3Q3FFD4
Ontologies
GO
PANTHER
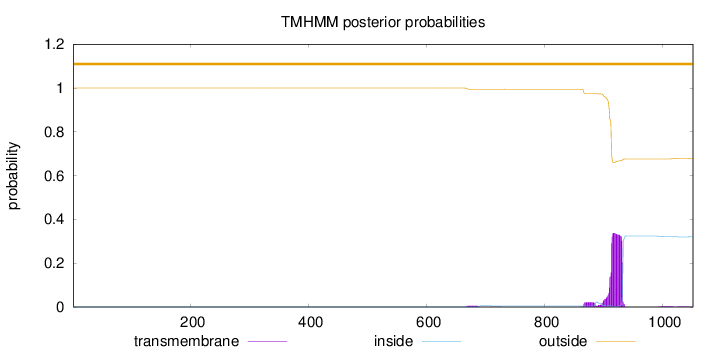
Topology
Length:
1051
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.15260999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00006
outside
1 - 1051
Population Genetic Test Statistics
Pi
234.717384
Theta
179.619297
Tajima's D
0.986124
CLR
0.004367
CSRT
0.656667166641668
Interpretation
Uncertain
