Gene
KWMTBOMO05635
Pre Gene Modal
BGIBMGA006783
Annotation
tRNA_(guanine-N(7)-)-methyltransferase_[Bombyx_mori]
Full name
tRNA (guanine-N(7)-)-methyltransferase
+ More
tRNA (guanine(46)-N(7))-methyltransferase
tRNA (guanine(46)-N(7))-methyltransferase
Alternative Name
tRNA (guanine(46)-N(7))-methyltransferase
tRNA(m7G46)-methyltransferase
Methyltransferase-like protein 1
tRNA(m7G46)-methyltransferase
Methyltransferase-like protein 1
Location in the cell
Cytoplasmic Reliability : 2.838
Sequence
CDS
ATGGCTTTGCCGCAAAAGAAATATTATAGACAACGTGCGCATTCTAACCCTATCGCAGATCACTGTTTTGATTATCCATCTCACCCTGATGATTATGACTGGACTCCCTTATATCCAATTATATCAGAAAAACCAAATCAGGTGGAGTTTTTAGATGTTGGTTGTGGTTATGGAGGACTTTTGGTTGCATTGTCTCCAATGTTTCCCAGTAACTTGATGTTGGGTTTGGAAATAAGAGTTAAAGTATCTGACTATGTAAATGATAGGATAAAAGCACTCAGGGTACAGTATCCTGATCAATATCAAAATGTAGCTGTGTTAAGGACAAATGCCATGAAATATCTTCCTAACTTTTTTCATAAAGGCCAGCTAAAAAAAATGTTTTTCTTATATCCAGATCCACATTTCAAGAAAGCAAAACATAAATGGAGGATAATTAACAAATGGCTCCTTTCTGAATATGCTTACGTACTGTGTGAACAGGGTATTGTATACACAATTACTGATGTAAAGGATTTGAATGAATGGATGGTTGCACATTTTGAGGAACATCCATTATTTGAAAGTGTTCCTGAAGAAGAATTGAAAGAAGATCCGATAATAGAAAAACTTTATGAGAGTACTGAAGAAGGTCAAAAAGTAACTAGGAATAATGGAGAGAAATTTCTAGCAGTGTTTAGAAGGATTGCTGATAAAACGAAAACTAATTAA
Protein
MALPQKKYYRQRAHSNPIADHCFDYPSHPDDYDWTPLYPIISEKPNQVEFLDVGCGYGGLLVALSPMFPSNLMLGLEIRVKVSDYVNDRIKALRVQYPDQYQNVAVLRTNAMKYLPNFFHKGQLKKMFFLYPDPHFKKAKHKWRIINKWLLSEYAYVLCEQGIVYTITDVKDLNEWMVAHFEEHPLFESVPEEELKEDPIIEKLYESTEEGQKVTRNNGEKFLAVFRRIADKTKTN
Summary
Description
Catalyzes the formation of N(7)-methylguanine at position 46 (m7G46) in tRNA.
Catalytic Activity
guanosine(46) in tRNA + S-adenosyl-L-methionine = N(7)-methylguanosine(46) in tRNA + S-adenosyl-L-homocysteine
Subunit
Forms a complex with WDR4.
Forms a complex with wdr4.
Forms a complex with wdr4.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. TrmB family.
Keywords
Complete proteome
Methyltransferase
Nucleus
Reference proteome
RNA-binding
S-adenosyl-L-methionine
Transferase
tRNA processing
tRNA-binding
Feature
chain tRNA (guanine-N(7)-)-methyltransferase
Uniprot
Q1HPU2
A0A2H1VV07
A0A2A4J8W6
A0A212EUR2
A0A3S2P042
A0A194QKT7
+ More
S4PBF3 A0A194PFS2 A0A026WGF8 J3JTL7 A0A232F597 A0A3L8E3Z7 A0A0J7KZC4 E2AU15 A0A1Y1K937 A0A2A3E4X7 F4WIR9 A0A154P1F2 A0A023GJK9 A0A1Y1K433 A0A023GH46 G3MQC9 A0A151JRN7 A0A195BNS6 A0A158P479 A0A151WLQ4 A0A310S9F9 E2B4E9 A0A1B6MQR3 A0A195CVC4 A0A131XD58 A0A1B6FR27 A0A0C9PIN7 A0A224Z2C4 A0A0C9QYQ8 L7M2C0 A0A293LHY9 A0A0M9ABR8 A0A2R5LBH8 A0A2S2NCG7 A0A2H8TS48 C3ZAN8 A0A195FNY8 E9FWC0 J9JNT7 A0A0V0G8T3 V4C725 A0A1S3HAD5 A0A0P5FS87 A0A0A9YJL3 A0A0P5CEL3 A0A224XWQ9 A0A146L0I9 A0A0P6HBG5 A0A0P5EHF5 A0A069DRA6 A0A034VG49 A0A2B4R9Z6 A0A1E1XBW4 Q0IEN3 A0A0P4XKE9 A0A2K7P6B8 A0A2P2HWG0 A0A0L8G1E2 C1BMF1 A0A1W4VEQ2 B3P8V8 A0A1I8MCP4 B4R338 R4G831 G1KN86 B4Q1B6 A0A182GV25 W8BEE3 A0A0P5RFW7 A0A1W4WLN1 B4I9N7 A0A084WQV1 A0A1I8NNL1 A0A182IRH4 A0A1A9VEK8 D3TR10 C1BTP4 A0A0M4EXW1 A0A1E1XR30 D3PHV0 A0A088AHP7 A0A3B4Z052 O77263 Q29I16 U5EUQ6 A0A2C9JZH1 B4N278 A0A1S4EL54 B4H4I3 A0A1L8HI54 Q6NU94 B4L529 A0A3Q1EN61
S4PBF3 A0A194PFS2 A0A026WGF8 J3JTL7 A0A232F597 A0A3L8E3Z7 A0A0J7KZC4 E2AU15 A0A1Y1K937 A0A2A3E4X7 F4WIR9 A0A154P1F2 A0A023GJK9 A0A1Y1K433 A0A023GH46 G3MQC9 A0A151JRN7 A0A195BNS6 A0A158P479 A0A151WLQ4 A0A310S9F9 E2B4E9 A0A1B6MQR3 A0A195CVC4 A0A131XD58 A0A1B6FR27 A0A0C9PIN7 A0A224Z2C4 A0A0C9QYQ8 L7M2C0 A0A293LHY9 A0A0M9ABR8 A0A2R5LBH8 A0A2S2NCG7 A0A2H8TS48 C3ZAN8 A0A195FNY8 E9FWC0 J9JNT7 A0A0V0G8T3 V4C725 A0A1S3HAD5 A0A0P5FS87 A0A0A9YJL3 A0A0P5CEL3 A0A224XWQ9 A0A146L0I9 A0A0P6HBG5 A0A0P5EHF5 A0A069DRA6 A0A034VG49 A0A2B4R9Z6 A0A1E1XBW4 Q0IEN3 A0A0P4XKE9 A0A2K7P6B8 A0A2P2HWG0 A0A0L8G1E2 C1BMF1 A0A1W4VEQ2 B3P8V8 A0A1I8MCP4 B4R338 R4G831 G1KN86 B4Q1B6 A0A182GV25 W8BEE3 A0A0P5RFW7 A0A1W4WLN1 B4I9N7 A0A084WQV1 A0A1I8NNL1 A0A182IRH4 A0A1A9VEK8 D3TR10 C1BTP4 A0A0M4EXW1 A0A1E1XR30 D3PHV0 A0A088AHP7 A0A3B4Z052 O77263 Q29I16 U5EUQ6 A0A2C9JZH1 B4N278 A0A1S4EL54 B4H4I3 A0A1L8HI54 Q6NU94 B4L529 A0A3Q1EN61
EC Number
2.1.1.33
Pubmed
22118469
26354079
23622113
24508170
22516182
28648823
+ More
30249741 20798317 28004739 21719571 22216098 21347285 28049606 28797301 25576852 18563158 21292972 23254933 25401762 26823975 26334808 25348373 28503490 17510324 17994087 25315136 26483478 24495485 24438588 20353571 29209593 10731132 12537572 10731137 12537569 15632085 15562597 27762356
30249741 20798317 28004739 21719571 22216098 21347285 28049606 28797301 25576852 18563158 21292972 23254933 25401762 26823975 26334808 25348373 28503490 17510324 17994087 25315136 26483478 24495485 24438588 20353571 29209593 10731132 12537572 10731137 12537569 15632085 15562597 27762356
EMBL
DQ443310
ODYU01004435
SOQ44302.1
NWSH01002591
PCG67900.1
AGBW02012332
+ More
OWR45201.1 RSAL01000073 RVE48977.1 KQ461198 KPJ06142.1 GAIX01005937 JAA86623.1 KQ459606 KPI91549.1 KK107260 EZA54144.1 BT126574 AEE61538.1 NNAY01000908 OXU25974.1 QOIP01000001 RLU26708.1 LBMM01001804 KMQ95716.1 GL442767 EFN63058.1 GEZM01093600 JAV56225.1 KZ288371 PBC26745.1 GL888177 EGI65865.1 KQ434796 KZC05652.1 GBBM01002195 JAC33223.1 GEZM01093601 JAV56224.1 GBBM01002224 JAC33194.1 JO844080 AEO35697.1 KQ978615 KYN29860.1 KQ976435 KYM87024.1 ADTU01008699 KQ982959 KYQ48721.1 KQ768478 OAD53131.1 GL445552 EFN89443.1 GEBQ01001707 JAT38270.1 KQ977279 KYN04119.1 GEFH01004239 JAP64342.1 GECZ01017103 JAS52666.1 GBYB01000803 JAG70570.1 GFPF01012860 MAA24006.1 GBYB01008944 JAG78711.1 GACK01007756 JAA57278.1 GFWV01003394 MAA28124.1 KQ435689 KOX81230.1 GGLE01002700 MBY06826.1 GGMR01002285 MBY14904.1 GFXV01005212 MBW17017.1 GG666602 EEN50441.1 KQ981382 KYN42275.1 GL732526 EFX88456.1 ABLF02023335 GECL01001628 JAP04496.1 KB201305 ESO97474.1 GDIP01144407 JAJ78995.1 GBHO01010347 GBRD01007047 JAG33257.1 JAG58774.1 GDIP01187552 JAJ35850.1 GFTR01004015 JAW12411.1 GDHC01016525 JAQ02104.1 GDIQ01022072 JAN72665.1 GDIP01159959 JAJ63443.1 GBGD01002707 JAC86182.1 GAKP01018409 JAC40543.1 LSMT01000777 PFX14461.1 GFAC01002443 JAT96745.1 CH477558 GDIP01241350 JAI82051.1 IACF01000323 LAB66111.1 KQ424537 KOF70846.1 BT075780 ACO10204.1 CH954183 CM000366 ACPB03018200 GAHY01001682 JAA75828.1 CM000162 JXUM01090180 KQ563823 KXJ73224.1 GAMC01009528 JAB97027.1 GDIQ01107589 GDIP01019965 JAL44137.1 JAM83750.1 CH480825 ATLV01025759 KE525399 KFB52595.1 EZ423862 ADD20138.1 BT077973 ACO12397.1 CP012528 ALC48692.1 GFAA01001644 JAU01791.1 BT121206 HACA01027719 ADD38136.1 CDW45080.1 AE014298 AL031765 AY094742 CH379064 GANO01001362 JAB58509.1 CH963925 CH479209 CM004468 OCT95780.1 BC068703 CH933811
OWR45201.1 RSAL01000073 RVE48977.1 KQ461198 KPJ06142.1 GAIX01005937 JAA86623.1 KQ459606 KPI91549.1 KK107260 EZA54144.1 BT126574 AEE61538.1 NNAY01000908 OXU25974.1 QOIP01000001 RLU26708.1 LBMM01001804 KMQ95716.1 GL442767 EFN63058.1 GEZM01093600 JAV56225.1 KZ288371 PBC26745.1 GL888177 EGI65865.1 KQ434796 KZC05652.1 GBBM01002195 JAC33223.1 GEZM01093601 JAV56224.1 GBBM01002224 JAC33194.1 JO844080 AEO35697.1 KQ978615 KYN29860.1 KQ976435 KYM87024.1 ADTU01008699 KQ982959 KYQ48721.1 KQ768478 OAD53131.1 GL445552 EFN89443.1 GEBQ01001707 JAT38270.1 KQ977279 KYN04119.1 GEFH01004239 JAP64342.1 GECZ01017103 JAS52666.1 GBYB01000803 JAG70570.1 GFPF01012860 MAA24006.1 GBYB01008944 JAG78711.1 GACK01007756 JAA57278.1 GFWV01003394 MAA28124.1 KQ435689 KOX81230.1 GGLE01002700 MBY06826.1 GGMR01002285 MBY14904.1 GFXV01005212 MBW17017.1 GG666602 EEN50441.1 KQ981382 KYN42275.1 GL732526 EFX88456.1 ABLF02023335 GECL01001628 JAP04496.1 KB201305 ESO97474.1 GDIP01144407 JAJ78995.1 GBHO01010347 GBRD01007047 JAG33257.1 JAG58774.1 GDIP01187552 JAJ35850.1 GFTR01004015 JAW12411.1 GDHC01016525 JAQ02104.1 GDIQ01022072 JAN72665.1 GDIP01159959 JAJ63443.1 GBGD01002707 JAC86182.1 GAKP01018409 JAC40543.1 LSMT01000777 PFX14461.1 GFAC01002443 JAT96745.1 CH477558 GDIP01241350 JAI82051.1 IACF01000323 LAB66111.1 KQ424537 KOF70846.1 BT075780 ACO10204.1 CH954183 CM000366 ACPB03018200 GAHY01001682 JAA75828.1 CM000162 JXUM01090180 KQ563823 KXJ73224.1 GAMC01009528 JAB97027.1 GDIQ01107589 GDIP01019965 JAL44137.1 JAM83750.1 CH480825 ATLV01025759 KE525399 KFB52595.1 EZ423862 ADD20138.1 BT077973 ACO12397.1 CP012528 ALC48692.1 GFAA01001644 JAU01791.1 BT121206 HACA01027719 ADD38136.1 CDW45080.1 AE014298 AL031765 AY094742 CH379064 GANO01001362 JAB58509.1 CH963925 CH479209 CM004468 OCT95780.1 BC068703 CH933811
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000053097 UP000215335 UP000279307 UP000036403 UP000000311 UP000242457 UP000007755 UP000076502 UP000078492 UP000078540 UP000005205 UP000075809 UP000008237 UP000078542 UP000053105 UP000001554 UP000078541 UP000000305 UP000007819 UP000030746 UP000085678 UP000225706 UP000008820 UP000053454 UP000192221 UP000008711 UP000095301 UP000000304 UP000015103 UP000001646 UP000002282 UP000069940 UP000249989 UP000192223 UP000001292 UP000030765 UP000095300 UP000075880 UP000078200 UP000092553 UP000005203 UP000261400 UP000000803 UP000001819 UP000076420 UP000007798 UP000079169 UP000008744 UP000186698 UP000009192 UP000257200
UP000053097 UP000215335 UP000279307 UP000036403 UP000000311 UP000242457 UP000007755 UP000076502 UP000078492 UP000078540 UP000005205 UP000075809 UP000008237 UP000078542 UP000053105 UP000001554 UP000078541 UP000000305 UP000007819 UP000030746 UP000085678 UP000225706 UP000008820 UP000053454 UP000192221 UP000008711 UP000095301 UP000000304 UP000015103 UP000001646 UP000002282 UP000069940 UP000249989 UP000192223 UP000001292 UP000030765 UP000095300 UP000075880 UP000078200 UP000092553 UP000005203 UP000261400 UP000000803 UP000001819 UP000076420 UP000007798 UP000079169 UP000008744 UP000186698 UP000009192 UP000257200
Pfam
PF02390 Methyltransf_4
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
Q1HPU2
A0A2H1VV07
A0A2A4J8W6
A0A212EUR2
A0A3S2P042
A0A194QKT7
+ More
S4PBF3 A0A194PFS2 A0A026WGF8 J3JTL7 A0A232F597 A0A3L8E3Z7 A0A0J7KZC4 E2AU15 A0A1Y1K937 A0A2A3E4X7 F4WIR9 A0A154P1F2 A0A023GJK9 A0A1Y1K433 A0A023GH46 G3MQC9 A0A151JRN7 A0A195BNS6 A0A158P479 A0A151WLQ4 A0A310S9F9 E2B4E9 A0A1B6MQR3 A0A195CVC4 A0A131XD58 A0A1B6FR27 A0A0C9PIN7 A0A224Z2C4 A0A0C9QYQ8 L7M2C0 A0A293LHY9 A0A0M9ABR8 A0A2R5LBH8 A0A2S2NCG7 A0A2H8TS48 C3ZAN8 A0A195FNY8 E9FWC0 J9JNT7 A0A0V0G8T3 V4C725 A0A1S3HAD5 A0A0P5FS87 A0A0A9YJL3 A0A0P5CEL3 A0A224XWQ9 A0A146L0I9 A0A0P6HBG5 A0A0P5EHF5 A0A069DRA6 A0A034VG49 A0A2B4R9Z6 A0A1E1XBW4 Q0IEN3 A0A0P4XKE9 A0A2K7P6B8 A0A2P2HWG0 A0A0L8G1E2 C1BMF1 A0A1W4VEQ2 B3P8V8 A0A1I8MCP4 B4R338 R4G831 G1KN86 B4Q1B6 A0A182GV25 W8BEE3 A0A0P5RFW7 A0A1W4WLN1 B4I9N7 A0A084WQV1 A0A1I8NNL1 A0A182IRH4 A0A1A9VEK8 D3TR10 C1BTP4 A0A0M4EXW1 A0A1E1XR30 D3PHV0 A0A088AHP7 A0A3B4Z052 O77263 Q29I16 U5EUQ6 A0A2C9JZH1 B4N278 A0A1S4EL54 B4H4I3 A0A1L8HI54 Q6NU94 B4L529 A0A3Q1EN61
S4PBF3 A0A194PFS2 A0A026WGF8 J3JTL7 A0A232F597 A0A3L8E3Z7 A0A0J7KZC4 E2AU15 A0A1Y1K937 A0A2A3E4X7 F4WIR9 A0A154P1F2 A0A023GJK9 A0A1Y1K433 A0A023GH46 G3MQC9 A0A151JRN7 A0A195BNS6 A0A158P479 A0A151WLQ4 A0A310S9F9 E2B4E9 A0A1B6MQR3 A0A195CVC4 A0A131XD58 A0A1B6FR27 A0A0C9PIN7 A0A224Z2C4 A0A0C9QYQ8 L7M2C0 A0A293LHY9 A0A0M9ABR8 A0A2R5LBH8 A0A2S2NCG7 A0A2H8TS48 C3ZAN8 A0A195FNY8 E9FWC0 J9JNT7 A0A0V0G8T3 V4C725 A0A1S3HAD5 A0A0P5FS87 A0A0A9YJL3 A0A0P5CEL3 A0A224XWQ9 A0A146L0I9 A0A0P6HBG5 A0A0P5EHF5 A0A069DRA6 A0A034VG49 A0A2B4R9Z6 A0A1E1XBW4 Q0IEN3 A0A0P4XKE9 A0A2K7P6B8 A0A2P2HWG0 A0A0L8G1E2 C1BMF1 A0A1W4VEQ2 B3P8V8 A0A1I8MCP4 B4R338 R4G831 G1KN86 B4Q1B6 A0A182GV25 W8BEE3 A0A0P5RFW7 A0A1W4WLN1 B4I9N7 A0A084WQV1 A0A1I8NNL1 A0A182IRH4 A0A1A9VEK8 D3TR10 C1BTP4 A0A0M4EXW1 A0A1E1XR30 D3PHV0 A0A088AHP7 A0A3B4Z052 O77263 Q29I16 U5EUQ6 A0A2C9JZH1 B4N278 A0A1S4EL54 B4H4I3 A0A1L8HI54 Q6NU94 B4L529 A0A3Q1EN61
PDB
3CKK
E-value=7.13962e-76,
Score=719
Ontologies
GO
PANTHER
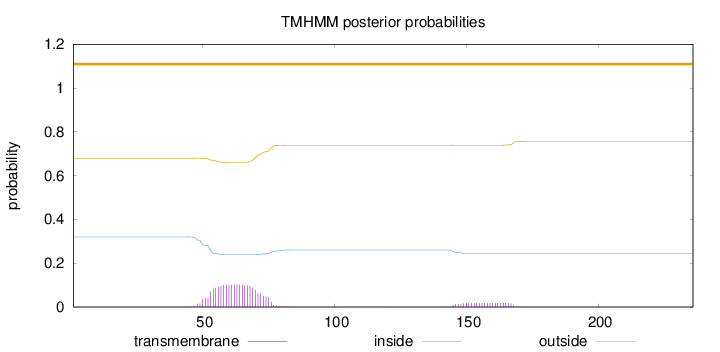
Topology
Subcellular location
Nucleus
Length:
236
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.57644
Exp number, first 60 AAs:
0.89252
Total prob of N-in:
0.32016
outside
1 - 236
Population Genetic Test Statistics
Pi
260.235528
Theta
186.323813
Tajima's D
1.136754
CLR
0.716855
CSRT
0.700664966751662
Interpretation
Uncertain
