Gene
KWMTBOMO05633
Pre Gene Modal
BGIBMGA006782
Annotation
hypothetical_protein_KGM_21825_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.273 Nuclear Reliability : 1.442
Sequence
CDS
ATGTCTGCAGTGGGCGAACCTGTGCTTAATATAATTGTTGTAGGCTTTCATCACAAGAAGGGATGTCAGGTGGAACATTGTTATCCTGAGGTTATACCTGGTCATCCGAATGAGCTACCAGCAAGTTGGCGACATTTGCCTGCTCTAGCTTTGCCTGATGGCTCTCACAATTACCATTCGGACACAATATTTTTTAATTTACCCGGACTCACAGAACCTGCTCACACTGTATATGGAATTTCATGTTTTAGACAAATCCCTATTGAGCAAGTGACTCAAAAGACAGAGGATATGACTAGAAGTTCAGTACAAAAGAGTGTTTGTGTAATATGTCGAGCTCCATTATTCGGTCGGCTTGCCGTGAAGATGGAGCTGGTAGTGAGGGCATGGTTTCTACAAGGTGACTTTTCACAGACAACACTCCTTGAAGATGCATACAAACATTTAAATAGTTGTCCTATTCAAACAGATCAAATTCTTGAAGGTTTATCAGTACAGCGATTGGTTGAAAACTGGAGGCACAAAGCATTGTTGTTATTTAAACTTCTGTTAATAAAACGTAAGGTTCTAATTTATGGATCACCAGCTGGCCCATTGTCAACTGTATTACTTACACTTGTGTCATTGCTTCCAAAATGTATAGAGAGTGGCCTTTCAAAAGCTGCAAATGTAGTATTATCAAGACCTTTATCTCCAATACCTATTTTAATAGATAATAAAATAGAAGATGCCATTGATGATAGCTGTGAAATTTTTCCTGAATCACTATTAAATGGTTCAAGTCAAGTAGAGGAATTAACACCTAAAGAATCTGGAAATATTTTGGATGAAAAAGACAGGGTCAGTAGACAAAGTTTTGATGAGGCTATCTTAACAGATGTGACCGATAGACAAGAATTTTCTACCAGAGATAAGTGCCATAGTATTGGTGAAAAATACAAGGTTCAAAAACCTGTGATAGATGCACAGCAAAGTCCCACTATGGCTAGAGACATGAGCGTTGATGGATTACACAGTTTAACTGGTCAAATTGATCAAGAAGAATGTGGATTTCCATTTCCACTGTTTGAAGATGGCTATCTATGTTTACCGTACTTATCATTGCAGTACTTAGACATTTTATCAGATCCTGCTGTTCATGGTTTCGTTGTCGGTGCATCTAATGTATTATTTAAGCAGAAGAGGCAGCTTTTTGATGTACTTGTAGAGTTAAATGAAATGAGAATAGAAACGGCAGATATGTTGCTCAGAAGGCAGTTGGCTCTTGGAACTGAGGATCTCAGATTTGCTGATCACGTTGTTCGTCATGGCGCAACTCAGGGCGATGCATGGATTAGAGATCAATTTGCAAGTTACTTGATATATATGTTAAGGACATCTATTTTGCCAGAAGGTAGTAGAGAGATAGACATGTTCAATGCTCAATATATGGCTGCTTTTAAGTCTACACCGGGCTACGAAAAGTGGTTGAAGAATACTAATAATGGTGATGTCGAAGCTTTCATGAACCTTATTCCTATGCATCCATTTTCTGGCCAACTCTCTGTAGCTGATATGAAGTTGAAGTTGGCACATACGATGTCGACGACGGAAGGCGGCAGGAAGGTGACGGCCGCGGTGGCGAGCACGTCCCGCGCGGTGGGCGGCGCGCTCTCGCAGGCGCGCGGCGCCCTCTCGGGCTGGTGGAGTGCGCTAACTGCCCCCCCGCCGCCACCGCCCCCGCCCCCCTCACCGCCGACCTCGCTCACTCCCGACCCGCCCGACGACCCCACAGACGCTACTGACATCACTGTGGTTTGA
Protein
MSAVGEPVLNIIVVGFHHKKGCQVEHCYPEVIPGHPNELPASWRHLPALALPDGSHNYHSDTIFFNLPGLTEPAHTVYGISCFRQIPIEQVTQKTEDMTRSSVQKSVCVICRAPLFGRLAVKMELVVRAWFLQGDFSQTTLLEDAYKHLNSCPIQTDQILEGLSVQRLVENWRHKALLLFKLLLIKRKVLIYGSPAGPLSTVLLTLVSLLPKCIESGLSKAANVVLSRPLSPIPILIDNKIEDAIDDSCEIFPESLLNGSSQVEELTPKESGNILDEKDRVSRQSFDEAILTDVTDRQEFSTRDKCHSIGEKYKVQKPVIDAQQSPTMARDMSVDGLHSLTGQIDQEECGFPFPLFEDGYLCLPYLSLQYLDILSDPAVHGFVVGASNVLFKQKRQLFDVLVELNEMRIETADMLLRRQLALGTEDLRFADHVVRHGATQGDAWIRDQFASYLIYMLRTSILPEGSREIDMFNAQYMAAFKSTPGYEKWLKNTNNGDVEAFMNLIPMHPFSGQLSVADMKLKLAHTMSTTEGGRKVTAAVASTSRAVGGALSQARGALSGWWSALTAPPPPPPPPPSPPTSLTPDPPDDPTDATDITVV
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
H9JB87
A0A2H1WLB1
A0A2A4JW46
A0A212EUN7
A0A194QKR0
A0A194PDY9
+ More
A0A1E1WIJ1 N6SZ13 A0A151WHV2 A0A195E801 A0A0C9RGK3 A0A158NF32 F4WEK7 A0A195AVI1 A0A088AGM7 E9J358 A0A195C639 A0A0M8ZX92 T1HHG6 E2BA71 A0A195FMA2 A0A1W4WMG6 A0A1W4WB59 U4UCN5 A0A0J7NZI4 A0A2A3E9C9 E2AE12 A0A0L7RDS7 A0A026WA97 A0A1B6KUY1 K7INH9 A0A232ERA0 A0A023F2U7 A0A146LQU0 A0A0A9XSC0 A0A139W954 A0A1Q3FWU8 A0A182PT82 Q7QF11 A0A1Q3FWN8 J9JUQ3 A0A2J7PME0 A0A084VH11 A0A182XY29 A0A139W991 A0A2H8TLP2 B3MVW2 A0A182VDZ8 A0A182W644 W5JRV3 A0A069DWM6 B4L4J7 A0A1B0G5M5 Q9VYB2 A0A182M752 B4Q2D5 A0A1A9ZBB5 A0A1A9UHY0 B4JJJ4 B4M2K9 A0A1S4F395 A0A2H8TZN9 A0A1A9Y8N0 Q17GP2 A0A182IAH6 A0A0Q9WCV0 A0A182FMD9 A0A182JDJ1 B4NUE2 A0A182H7E4 B3NWM0 A0A1A9W939 A0A3B0JWT6 A0A182N0C5 A0A1L8DAQ7 A0A0A1XJH5 A0A182QJ42 A0A1I8QDT4 A0A1W4WDS4 B4GXM9 Q29HH8 A0A0K8UDK5 A0A154PU00 B4MSK8 A0A1L8DAM1 A0A182TT15 A0A1B0C3V6 A0A182G2Q8 A0A0L0C4V4 A0A0T6B479 E9FU23 A0A0P6IIQ0 A0A0P5ZQ08 A0A226D226 A0A336KFQ2 A0A034VCN5 A0A0P6FPH6 A0A0P5U210 A0A0N8DPF3
A0A1E1WIJ1 N6SZ13 A0A151WHV2 A0A195E801 A0A0C9RGK3 A0A158NF32 F4WEK7 A0A195AVI1 A0A088AGM7 E9J358 A0A195C639 A0A0M8ZX92 T1HHG6 E2BA71 A0A195FMA2 A0A1W4WMG6 A0A1W4WB59 U4UCN5 A0A0J7NZI4 A0A2A3E9C9 E2AE12 A0A0L7RDS7 A0A026WA97 A0A1B6KUY1 K7INH9 A0A232ERA0 A0A023F2U7 A0A146LQU0 A0A0A9XSC0 A0A139W954 A0A1Q3FWU8 A0A182PT82 Q7QF11 A0A1Q3FWN8 J9JUQ3 A0A2J7PME0 A0A084VH11 A0A182XY29 A0A139W991 A0A2H8TLP2 B3MVW2 A0A182VDZ8 A0A182W644 W5JRV3 A0A069DWM6 B4L4J7 A0A1B0G5M5 Q9VYB2 A0A182M752 B4Q2D5 A0A1A9ZBB5 A0A1A9UHY0 B4JJJ4 B4M2K9 A0A1S4F395 A0A2H8TZN9 A0A1A9Y8N0 Q17GP2 A0A182IAH6 A0A0Q9WCV0 A0A182FMD9 A0A182JDJ1 B4NUE2 A0A182H7E4 B3NWM0 A0A1A9W939 A0A3B0JWT6 A0A182N0C5 A0A1L8DAQ7 A0A0A1XJH5 A0A182QJ42 A0A1I8QDT4 A0A1W4WDS4 B4GXM9 Q29HH8 A0A0K8UDK5 A0A154PU00 B4MSK8 A0A1L8DAM1 A0A182TT15 A0A1B0C3V6 A0A182G2Q8 A0A0L0C4V4 A0A0T6B479 E9FU23 A0A0P6IIQ0 A0A0P5ZQ08 A0A226D226 A0A336KFQ2 A0A034VCN5 A0A0P6FPH6 A0A0P5U210 A0A0N8DPF3
Pubmed
19121390
22118469
26354079
23537049
21347285
21719571
+ More
21282665 20798317 24508170 30249741 20075255 28648823 25474469 26823975 25401762 18362917 19820115 12364791 14747013 17210077 24438588 25244985 17994087 20920257 23761445 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 18057021 26483478 25830018 15632085 26108605 21292972 25348373
21282665 20798317 24508170 30249741 20075255 28648823 25474469 26823975 25401762 18362917 19820115 12364791 14747013 17210077 24438588 25244985 17994087 20920257 23761445 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 18057021 26483478 25830018 15632085 26108605 21292972 25348373
EMBL
BABH01008780
ODYU01009430
SOQ53849.1
NWSH01000460
PCG76245.1
AGBW02012332
+ More
OWR45205.1 KQ461198 KPJ06148.1 KQ459606 KPI91556.1 GDQN01004286 JAT86768.1 APGK01059304 KB741292 ENN70483.1 KQ983106 KYQ47397.1 KQ979479 KYN21345.1 GBYB01006161 JAG75928.1 ADTU01013722 ADTU01013723 GL888103 EGI67370.1 KQ976736 KYM76035.1 GL768057 EFZ12743.1 KQ978287 KYM95628.1 KQ435840 KOX71463.1 ACPB03009319 GL446678 EFN87413.1 KQ981490 KYN41372.1 KB632203 ERL90078.1 LBMM01000675 KMQ97810.1 KZ288344 PBC27661.1 GL438827 EFN68245.1 KQ414612 KOC69142.1 KK107295 QOIP01000009 EZA52975.1 RLU18761.1 GEBQ01024730 JAT15247.1 NNAY01002633 OXU20890.1 GBBI01003052 JAC15660.1 GDHC01008980 JAQ09649.1 GBHO01021896 GBRD01006686 JAG21708.1 JAG59135.1 KQ972669 KXZ75822.1 GFDL01003027 JAV32018.1 AAAB01008846 EAA06436.5 GFDL01003046 JAV31999.1 ABLF02024512 NEVH01024022 PNF17496.1 ATLV01013122 KE524840 KFB37255.1 KXZ75821.1 GFXV01003272 MBW15077.1 CH902625 EDV35107.1 ADMH02000649 ETN65459.1 GBGD01000782 JAC88107.1 CH933810 EDW07475.1 CCAG010013941 AE014298 AY061257 AAF48291.1 AAL28805.1 AXCM01001058 CM000162 EDX01596.1 CH916370 EDV99746.1 CH940651 EDW65913.1 GFXV01007416 MBW19221.1 CH477257 EAT45825.1 APCN01001239 KRF82475.1 KRF82476.1 CH983913 EDX16589.1 JXUM01116069 KQ565945 KXJ70507.1 CH954180 EDV47182.1 OUUW01000011 SPP86547.1 GFDF01010533 JAV03551.1 GBXI01003141 GBXI01001174 JAD11151.1 JAD13118.1 AXCN02001229 CH479196 EDW27506.1 CH379064 EAL31780.2 GDHF01027661 JAI24653.1 KQ435163 KZC14924.1 CH963851 EDW75097.1 GFDF01010602 JAV03482.1 JXJN01025121 JXUM01139898 JXUM01139899 KQ568972 KXJ68814.1 JRES01000984 KNC26474.1 LJIG01009892 KRT82172.1 GL732524 EFX89542.1 GDIQ01010408 JAN84329.1 GDIP01040800 LRGB01003123 JAM62915.1 KZS04515.1 LNIX01000042 OXA38924.1 UFQS01000315 UFQT01000315 SSX02742.1 SSX23114.1 GAKP01018716 GAKP01018713 JAC40239.1 GDIQ01044560 JAN50177.1 GDIP01120840 JAL82874.1 GDIP01013823 JAM89892.1
OWR45205.1 KQ461198 KPJ06148.1 KQ459606 KPI91556.1 GDQN01004286 JAT86768.1 APGK01059304 KB741292 ENN70483.1 KQ983106 KYQ47397.1 KQ979479 KYN21345.1 GBYB01006161 JAG75928.1 ADTU01013722 ADTU01013723 GL888103 EGI67370.1 KQ976736 KYM76035.1 GL768057 EFZ12743.1 KQ978287 KYM95628.1 KQ435840 KOX71463.1 ACPB03009319 GL446678 EFN87413.1 KQ981490 KYN41372.1 KB632203 ERL90078.1 LBMM01000675 KMQ97810.1 KZ288344 PBC27661.1 GL438827 EFN68245.1 KQ414612 KOC69142.1 KK107295 QOIP01000009 EZA52975.1 RLU18761.1 GEBQ01024730 JAT15247.1 NNAY01002633 OXU20890.1 GBBI01003052 JAC15660.1 GDHC01008980 JAQ09649.1 GBHO01021896 GBRD01006686 JAG21708.1 JAG59135.1 KQ972669 KXZ75822.1 GFDL01003027 JAV32018.1 AAAB01008846 EAA06436.5 GFDL01003046 JAV31999.1 ABLF02024512 NEVH01024022 PNF17496.1 ATLV01013122 KE524840 KFB37255.1 KXZ75821.1 GFXV01003272 MBW15077.1 CH902625 EDV35107.1 ADMH02000649 ETN65459.1 GBGD01000782 JAC88107.1 CH933810 EDW07475.1 CCAG010013941 AE014298 AY061257 AAF48291.1 AAL28805.1 AXCM01001058 CM000162 EDX01596.1 CH916370 EDV99746.1 CH940651 EDW65913.1 GFXV01007416 MBW19221.1 CH477257 EAT45825.1 APCN01001239 KRF82475.1 KRF82476.1 CH983913 EDX16589.1 JXUM01116069 KQ565945 KXJ70507.1 CH954180 EDV47182.1 OUUW01000011 SPP86547.1 GFDF01010533 JAV03551.1 GBXI01003141 GBXI01001174 JAD11151.1 JAD13118.1 AXCN02001229 CH479196 EDW27506.1 CH379064 EAL31780.2 GDHF01027661 JAI24653.1 KQ435163 KZC14924.1 CH963851 EDW75097.1 GFDF01010602 JAV03482.1 JXJN01025121 JXUM01139898 JXUM01139899 KQ568972 KXJ68814.1 JRES01000984 KNC26474.1 LJIG01009892 KRT82172.1 GL732524 EFX89542.1 GDIQ01010408 JAN84329.1 GDIP01040800 LRGB01003123 JAM62915.1 KZS04515.1 LNIX01000042 OXA38924.1 UFQS01000315 UFQT01000315 SSX02742.1 SSX23114.1 GAKP01018716 GAKP01018713 JAC40239.1 GDIQ01044560 JAN50177.1 GDIP01120840 JAL82874.1 GDIP01013823 JAM89892.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000019118
+ More
UP000075809 UP000078492 UP000005205 UP000007755 UP000078540 UP000005203 UP000078542 UP000053105 UP000015103 UP000008237 UP000078541 UP000192223 UP000030742 UP000036403 UP000242457 UP000000311 UP000053825 UP000053097 UP000279307 UP000002358 UP000215335 UP000007266 UP000075885 UP000007062 UP000007819 UP000235965 UP000030765 UP000076408 UP000007801 UP000075903 UP000075920 UP000000673 UP000009192 UP000092444 UP000000803 UP000075883 UP000002282 UP000092445 UP000078200 UP000001070 UP000008792 UP000092443 UP000008820 UP000075840 UP000069272 UP000075880 UP000000304 UP000069940 UP000249989 UP000008711 UP000091820 UP000268350 UP000075884 UP000075886 UP000095300 UP000192221 UP000008744 UP000001819 UP000076502 UP000007798 UP000075902 UP000092460 UP000037069 UP000000305 UP000076858 UP000198287
UP000075809 UP000078492 UP000005205 UP000007755 UP000078540 UP000005203 UP000078542 UP000053105 UP000015103 UP000008237 UP000078541 UP000192223 UP000030742 UP000036403 UP000242457 UP000000311 UP000053825 UP000053097 UP000279307 UP000002358 UP000215335 UP000007266 UP000075885 UP000007062 UP000007819 UP000235965 UP000030765 UP000076408 UP000007801 UP000075903 UP000075920 UP000000673 UP000009192 UP000092444 UP000000803 UP000075883 UP000002282 UP000092445 UP000078200 UP000001070 UP000008792 UP000092443 UP000008820 UP000075840 UP000069272 UP000075880 UP000000304 UP000069940 UP000249989 UP000008711 UP000091820 UP000268350 UP000075884 UP000075886 UP000095300 UP000192221 UP000008744 UP000001819 UP000076502 UP000007798 UP000075902 UP000092460 UP000037069 UP000000305 UP000076858 UP000198287
Interpro
ProteinModelPortal
H9JB87
A0A2H1WLB1
A0A2A4JW46
A0A212EUN7
A0A194QKR0
A0A194PDY9
+ More
A0A1E1WIJ1 N6SZ13 A0A151WHV2 A0A195E801 A0A0C9RGK3 A0A158NF32 F4WEK7 A0A195AVI1 A0A088AGM7 E9J358 A0A195C639 A0A0M8ZX92 T1HHG6 E2BA71 A0A195FMA2 A0A1W4WMG6 A0A1W4WB59 U4UCN5 A0A0J7NZI4 A0A2A3E9C9 E2AE12 A0A0L7RDS7 A0A026WA97 A0A1B6KUY1 K7INH9 A0A232ERA0 A0A023F2U7 A0A146LQU0 A0A0A9XSC0 A0A139W954 A0A1Q3FWU8 A0A182PT82 Q7QF11 A0A1Q3FWN8 J9JUQ3 A0A2J7PME0 A0A084VH11 A0A182XY29 A0A139W991 A0A2H8TLP2 B3MVW2 A0A182VDZ8 A0A182W644 W5JRV3 A0A069DWM6 B4L4J7 A0A1B0G5M5 Q9VYB2 A0A182M752 B4Q2D5 A0A1A9ZBB5 A0A1A9UHY0 B4JJJ4 B4M2K9 A0A1S4F395 A0A2H8TZN9 A0A1A9Y8N0 Q17GP2 A0A182IAH6 A0A0Q9WCV0 A0A182FMD9 A0A182JDJ1 B4NUE2 A0A182H7E4 B3NWM0 A0A1A9W939 A0A3B0JWT6 A0A182N0C5 A0A1L8DAQ7 A0A0A1XJH5 A0A182QJ42 A0A1I8QDT4 A0A1W4WDS4 B4GXM9 Q29HH8 A0A0K8UDK5 A0A154PU00 B4MSK8 A0A1L8DAM1 A0A182TT15 A0A1B0C3V6 A0A182G2Q8 A0A0L0C4V4 A0A0T6B479 E9FU23 A0A0P6IIQ0 A0A0P5ZQ08 A0A226D226 A0A336KFQ2 A0A034VCN5 A0A0P6FPH6 A0A0P5U210 A0A0N8DPF3
A0A1E1WIJ1 N6SZ13 A0A151WHV2 A0A195E801 A0A0C9RGK3 A0A158NF32 F4WEK7 A0A195AVI1 A0A088AGM7 E9J358 A0A195C639 A0A0M8ZX92 T1HHG6 E2BA71 A0A195FMA2 A0A1W4WMG6 A0A1W4WB59 U4UCN5 A0A0J7NZI4 A0A2A3E9C9 E2AE12 A0A0L7RDS7 A0A026WA97 A0A1B6KUY1 K7INH9 A0A232ERA0 A0A023F2U7 A0A146LQU0 A0A0A9XSC0 A0A139W954 A0A1Q3FWU8 A0A182PT82 Q7QF11 A0A1Q3FWN8 J9JUQ3 A0A2J7PME0 A0A084VH11 A0A182XY29 A0A139W991 A0A2H8TLP2 B3MVW2 A0A182VDZ8 A0A182W644 W5JRV3 A0A069DWM6 B4L4J7 A0A1B0G5M5 Q9VYB2 A0A182M752 B4Q2D5 A0A1A9ZBB5 A0A1A9UHY0 B4JJJ4 B4M2K9 A0A1S4F395 A0A2H8TZN9 A0A1A9Y8N0 Q17GP2 A0A182IAH6 A0A0Q9WCV0 A0A182FMD9 A0A182JDJ1 B4NUE2 A0A182H7E4 B3NWM0 A0A1A9W939 A0A3B0JWT6 A0A182N0C5 A0A1L8DAQ7 A0A0A1XJH5 A0A182QJ42 A0A1I8QDT4 A0A1W4WDS4 B4GXM9 Q29HH8 A0A0K8UDK5 A0A154PU00 B4MSK8 A0A1L8DAM1 A0A182TT15 A0A1B0C3V6 A0A182G2Q8 A0A0L0C4V4 A0A0T6B479 E9FU23 A0A0P6IIQ0 A0A0P5ZQ08 A0A226D226 A0A336KFQ2 A0A034VCN5 A0A0P6FPH6 A0A0P5U210 A0A0N8DPF3
Ontologies
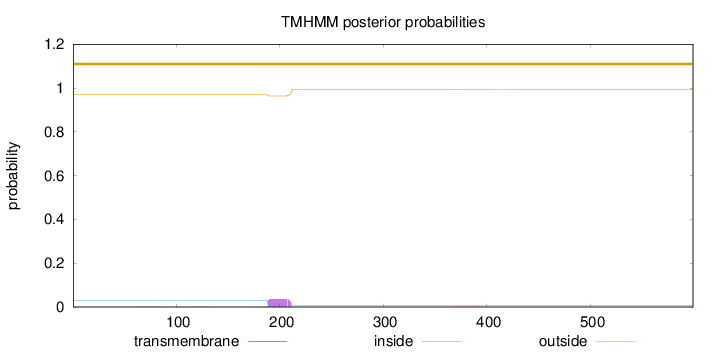
Topology
Length:
599
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.830029999999998
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.03030
outside
1 - 599
Population Genetic Test Statistics
Pi
274.397625
Theta
181.68492
Tajima's D
1.453155
CLR
0
CSRT
0.781560921953902
Interpretation
Uncertain
