Gene
KWMTBOMO05630
Pre Gene Modal
BGIBMGA006555
Annotation
PREDICTED:_Ca(2+)/calmodulin-responsive_adenylate_cyclase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.856 PlasmaMembrane Reliability : 1.46
Sequence
CDS
ATGGTTGGCGTATTGAGTATCCACGCTGTGCTGTGCAGCGGATTGGCGTGGTGGTGCGGCCGGACTGACGGTGTGGCACCAGAGAGGCGGTTGCCGCGGGCATGCTGGCTGGCCCTGGCTGGCGGGGGTGCGGTACTAGCTGCCACGCTGCCCGCCTGGAGCCCGGGTGCTGCAGCCGAGGGCGCTGTGCATGTCGTCTGGGCTGTCTTCGCTGCCTATGCTCTGCTACCTGTTGGCGCTCCTGTCGCTGCCGCCTTTGGCATTGTCTTACCAACAGTCCATACCATCGCCGCTGCGCTTGTTGCTCATCGCTTTCCTTATCACGTCTGGCAGCAGATGCTGGGCAATGTGGTCGTGTTCGTTTGTGTGAATGTGGTTGGAGCTTTGATGCACTCGCTGATGGAAACCGCCCAGCGCAGAGCCTTCCTAGACACCAGGAACTGCATCGCGGCGAGACTCGACATGGAAGACGAAAACGAGAAATTGGAAAGGTTACTGTTATCCGTTCTCCCGCAACACGTCGCTATGGAGATGAAGAACGATATCATATCGCCGGTCGAGGGACAGTTCCACAAGATATACATACAGAAACACGAACAAGTCAGCATACTGTTTGCGGACATCGTCGGTTTTACTGTACTCGCTTCGCAGTGCAGCGCGCAGGAGCTGGTGCGGCTACTGAACGAACTGTTCGGAAGATTCGATCAATTGGCGAACGACAATCATTGCCTCCGCATCAAGATCCTGGGTGACTGTTACTACTGCGTGTCGGGATTGCCGGAGCCGCGCTCCGACCACGCTCGCTGCACCGTCGAGATGGGACTAGACATGATCGATGGCATCGCGTCAGTTGTGGAGGCTACCGACGTACAGCTAAACATGCGGGTGGGAATACACACGGGCCGAGTGTTGTGCGGAGTGCTCGGTCTTCGCAAGTGGCAGTACGATGTGTGGTCCAATGACGTCACTCTTGCAAACAATATGGAAGCTGGCGGCGAACCTGGTCGTGTACACATCACCCAAGCCACGTTGGAGTGCCTGGGAGGTGCGTACGAGGTAGAGCCGGGCCACGGAGCTAGTCGCAACGCGTACCTAAGGGACCACTCTGTTCAGACATACTTCATTATACCGCCGCCCAGGAGAAGAAAAATGTGTTTATCTTCGACGGCGGGACTGGGCACGGGCTCGCGTCGGAAACTATCCTTCAAGAACGTTTCCAACGTGGTCGTACAACTCCTGCACTCGATTAAGTTCAACGTGGATGTTCCGTTCTCGAATATGGCAGCGTCGCCTCAGGAACTCAAGGCCAATGCAGCCCGGAAGAATAAGGTAACGGAGAAATTTAAGCGGCCTCTGAAAAAGCGTCACTCGTCTGTCTACCACCAGCCCACCAACCGCGTCAACAAGTACCTAGCGCAAGCCATAGACGCCCGTTCCGTGCATCGCGAGAAGGCAACTCACGTGCATCTGCTAACGTTGTGCTTCAAACAAGCAGACAAAGAGGCTCAGTACCGCGCGTCAACGGACGGCGGCTGGGCCGGGTCGCTGGGTGCGGCTCTGGGCGCGCTGGTGGCGGCGGGCACGCTGCAGGCCGCCATCTTGCCGCGCACCACCATCTTGTTGCTGCTCTTCGTCACCGCTTTCGCGTGGTCGGCTGCCGTTCTCGCGTTGACTCTAGCCGCTCGACTGAGGCTCATACCTTGCGATGTATCCAGGCCGTTCGCTCTCAGGAACGCCATCACGACCTTCACTCTCGTGCTGGGCTATGCCGTGGGCCAAGTTAATGTGGTGACGTGCCGCAAAGTGGACGTGTGTCGGAACCTCACCGAGAACGTAACGATGGATGATATCAGTATGGCGAGTGACGACGTCGCGTCTCTGCTGTGGAGCAGCGCCGACCACCGCGCGTGTCCGGTGCCGCTGTACGTGACGCTGGGCTGCTGCCTCAGCATGCTGGCGGTCGCGGGCTTCCTCAGACTGCCGATACTGATCAAGGGCATCCTCCTCTACGTCATGACGGCCGGGTACATGGTAGTCATACTGGCCTACCATAAGGACTTGTTCGACTGCTACGATCTAATGACCGAACCGGGGCCAGTTGGGTCAGCGTACGTCGCCTGCGCCTGGACGCTGGCACTGGCACTGGCAGTGTTATTGCACGCGCGGCAGACGGAGTGGACGGCGCGACTCGACTTCTTGTGGCAAGTTCAAGCGCGCGATGAGAAAAGAGACATGGACGCTCTACAGGCATCAAACAGAAGAATACTGTTCAACCTCTTACCGGCGCACGTGGCCACTCACTTCTTGGATAATCAATTTAGAACAAATATGGACCTGTACCACCAGTCGTACCAGCGCGTCGGCGTCGTGTTCGCCTCCATCACCAACTACCACGAGTTCTACATGGAGCTCGACGGGAACAACCAGGGCATGGAGTGTCTGCGCTTGCTCAACGAAATCATAGCCGACTTCGATGAGCTTTTAGGTGAAGATCGCTTCAGTGCAATAGATAAAATAAAGACTGTCGGCTCAACGTACATGGCTGCCGTCGGTTTGATCCCGGACAAAAAAATGGCGGACGAAGTAAGCACGCGCAAGCACATGGCCACCCTGGTTGAATTCGTTTTTTCTATGAGGGACAAACTCAAAGACATCAACGACAACTCATACAATAACTTTATGCTCAGAGTCGGTATCAACGTCGGGCCCGTGGTGGCCGGTGTGATTGGCGCGAGGAAACCACAGTACGACATTTGGGGGAACACCGTCAATGTTGCCTCACGAATGGATTCCACCGGCCTGCCGAATCACACTCAAGTTACAGAAGAAGTGTATCACGTGTTGAAGGATATGCCGTACCAGTTTGTGTGCAGGGGGAAGGTCAAAGTTAAAGGGAAGGGCGAGATGACTACTTACTTCTTGACGGACCGAGCGCCAACGAACGGCACTGCGCAGAACCCGGCTAATCCTCCATCTACGAATCCCGCGACCGCCTACGGCGGCGTGGCCACTCCGCTAGCGATGCTGCAGAACTCGGCCAGGAGGGCGGCGGCGCAGCCATCGAGACTGCCGCCGGTGCGAGAGGCCTCGGTCGCAGGCGAGAACGAACCTCTATTGCCCACTAACAGCCAACAGATCAACGGTAATAATCACAAGGGCAGTAAAGGTCGCAGGAACAAAGACTCTATCCCTGAAGAAGCGCCACCGCCGCCTCCTCCCCATGGAATGTCAAACCCTAGATGGCCGCCGCCTCGAGCACTGGTGCCGCCCTGGCCCCGTCCGCAGGAGCCCCGGCCTCCAGTAGCGCACAAGAGACGGCCCCCGACGCGCCTGCCGCGGCCTCGCTCTAGTGAGAGCCTACCGTTAACGCGGAGCCCCCGGGTCCATTCGTCTGCGGACGAGTTAAGTTCTTTAACGCGATCTCCGAGTCTTAGTTCGTCAGACGAAAGCTATTCGCGAACAACTGACGCTTCTCCTTCGCCTCCCCGCATATCTCAATGGCTCCCCGCCTCACGGCCGAACCGGTCGAGTCCTAATCAGCCGTACGACTACCCGCCTTCAAGAGCGACGAAGCCTGTTTCGAATCACATCAACGAAATATATTCGAAGCATAAAATATCAAAAAAACCATCGTTAGACGACAAGTTTTTAGATGTAGATCATCAGAAAGAGGATGAAAAACTTCCAAGGACCATCATTAATCTATTACAAACTTCGGCGAAGCGTGATGGGTGCTGTCAAACGGACAAGAAGGAAGGCCGCATGGGACAACGAGCCAGCTCATTCTCCAGTTCGGGCAGCAGACCGAATAATTTCAAACATTTCAATCACGATTCGGTAGACGTGTACACAAAAAGGAGTCCATCCGACGTGGCGTGCGTGCCGCCTTTCGAAAGGGAAATACAAAGACTATTAGAGGATAAAAATCTCATCAAAAGCAATCCACCCAAACTCGAAAGGAAAGAACTCAAACTCGATCTGAGGAGCCAGAACCCGTCGCTGGATTCGGTTCGCAACAATAACAACAACAACAACAGCAGCCCGCTGCACGCGGTGGGGCTGGCGGCCATCGCTCAGCTGGCGCGGCAGGACGCCCCCCCTGCCGCCCCTGCCGGTGACTTCCCCGGCGTGCTGCCCACCGACGTCGTCGTCCAGCTGCCCTCACCCACGCACTCAAGGGAACAACGCTCGACCCCGCAGTTTGAAAGGAAAGCGGAGGCCCTGGCGATGAAAGAAAAACACGAGAAAGAAATTCAAGAAAGACTACAAAGCAGGGTGACCACTTCTAACTTGAGGGAAATGGGCCTCTACGGCGAAAGTCAGTCTGAATGGAGTTCGTCCTGTTCGGAGGGAGAAGGCGACGGAGACTGTGGACACCCCGAGAGCACCGGATATACCACAGACGACCCCGCCCTCGAAAACGTGTCGATGCTCAACGAGGCCGGCCTCACGGACGCCGAGGCGGCCCTCAGCGACGTGAACTCCTGCTACCTGGACCGCGACGACGACGTCTCCTCGCGGGCCTCCTCGCGCCTCCTCGACTCCGAGGCGCTCGTGTCCCTGGAGTCGCTGAGCGCTATATACGACTCCGACGAACCCGCCCACAGGTACTTGGCCAACATCCGTACCGTCAGCGAATCCATTACGAGGAACTTCGGGCAGCCGGCGCACGTCACCGACGCCGAAAGCGATGTTTAG
Protein
MVGVLSIHAVLCSGLAWWCGRTDGVAPERRLPRACWLALAGGGAVLAATLPAWSPGAAAEGAVHVVWAVFAAYALLPVGAPVAAAFGIVLPTVHTIAAALVAHRFPYHVWQQMLGNVVVFVCVNVVGALMHSLMETAQRRAFLDTRNCIAARLDMEDENEKLERLLLSVLPQHVAMEMKNDIISPVEGQFHKIYIQKHEQVSILFADIVGFTVLASQCSAQELVRLLNELFGRFDQLANDNHCLRIKILGDCYYCVSGLPEPRSDHARCTVEMGLDMIDGIASVVEATDVQLNMRVGIHTGRVLCGVLGLRKWQYDVWSNDVTLANNMEAGGEPGRVHITQATLECLGGAYEVEPGHGASRNAYLRDHSVQTYFIIPPPRRRKMCLSSTAGLGTGSRRKLSFKNVSNVVVQLLHSIKFNVDVPFSNMAASPQELKANAARKNKVTEKFKRPLKKRHSSVYHQPTNRVNKYLAQAIDARSVHREKATHVHLLTLCFKQADKEAQYRASTDGGWAGSLGAALGALVAAGTLQAAILPRTTILLLLFVTAFAWSAAVLALTLAARLRLIPCDVSRPFALRNAITTFTLVLGYAVGQVNVVTCRKVDVCRNLTENVTMDDISMASDDVASLLWSSADHRACPVPLYVTLGCCLSMLAVAGFLRLPILIKGILLYVMTAGYMVVILAYHKDLFDCYDLMTEPGPVGSAYVACAWTLALALAVLLHARQTEWTARLDFLWQVQARDEKRDMDALQASNRRILFNLLPAHVATHFLDNQFRTNMDLYHQSYQRVGVVFASITNYHEFYMELDGNNQGMECLRLLNEIIADFDELLGEDRFSAIDKIKTVGSTYMAAVGLIPDKKMADEVSTRKHMATLVEFVFSMRDKLKDINDNSYNNFMLRVGINVGPVVAGVIGARKPQYDIWGNTVNVASRMDSTGLPNHTQVTEEVYHVLKDMPYQFVCRGKVKVKGKGEMTTYFLTDRAPTNGTAQNPANPPSTNPATAYGGVATPLAMLQNSARRAAAQPSRLPPVREASVAGENEPLLPTNSQQINGNNHKGSKGRRNKDSIPEEAPPPPPPHGMSNPRWPPPRALVPPWPRPQEPRPPVAHKRRPPTRLPRPRSSESLPLTRSPRVHSSADELSSLTRSPSLSSSDESYSRTTDASPSPPRISQWLPASRPNRSSPNQPYDYPPSRATKPVSNHINEIYSKHKISKKPSLDDKFLDVDHQKEDEKLPRTIINLLQTSAKRDGCCQTDKKEGRMGQRASSFSSSGSRPNNFKHFNHDSVDVYTKRSPSDVACVPPFEREIQRLLEDKNLIKSNPPKLERKELKLDLRSQNPSLDSVRNNNNNNNSSPLHAVGLAAIAQLARQDAPPAAPAGDFPGVLPTDVVVQLPSPTHSREQRSTPQFERKAEALAMKEKHEKEIQERLQSRVTTSNLREMGLYGESQSEWSSSCSEGEGDGDCGHPESTGYTTDDPALENVSMLNEAGLTDAEAALSDVNSCYLDRDDDVSSRASSRLLDSEALVSLESLSAIYDSDEPAHRYLANIRTVSESITRNFGQPAHVTDAESDV
Summary
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF55073
SSF55073
Gene 3D
ProteinModelPortal
PDB
3MAA
E-value=1.07753e-87,
Score=830
Ontologies
PATHWAY
GO
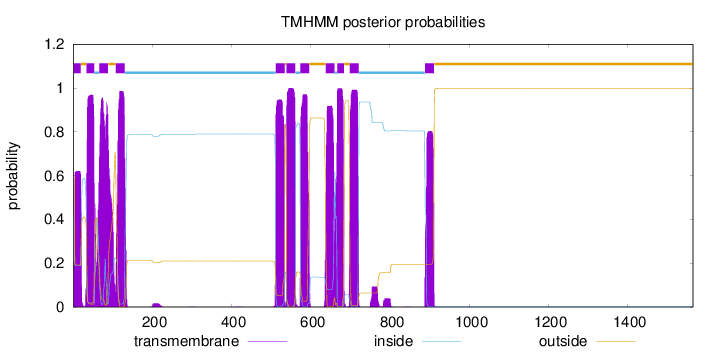
Topology
Length:
1565
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
231.05074
Exp number, first 60 AAs:
32.67422
Total prob of N-in:
0.40767
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 19
outside
20 - 33
TMhelix
34 - 53
inside
54 - 65
TMhelix
66 - 88
outside
89 - 107
TMhelix
108 - 130
inside
131 - 511
TMhelix
512 - 534
outside
535 - 538
TMhelix
539 - 561
inside
562 - 573
TMhelix
574 - 596
outside
597 - 637
TMhelix
638 - 660
inside
661 - 666
TMhelix
667 - 684
outside
685 - 698
TMhelix
699 - 721
inside
722 - 888
TMhelix
889 - 911
outside
912 - 1565
Population Genetic Test Statistics
Pi
238.898634
Theta
177.27726
Tajima's D
1.022098
CLR
0.239777
CSRT
0.667116644167792
Interpretation
Uncertain
