Gene
KWMTBOMO05626
Pre Gene Modal
BGIBMGA006559
Annotation
PREDICTED:_sn1-specific_diacylglycerol_lipase_beta_isoform_X2_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.508
Sequence
CDS
ATGGCCCGAATGCCGGCGCTCCGTCTTCTCGGACGCAAATGGCTGGCAGCCTCGGATGATCTCGTCTTCCCAAGCATTTTCGAATTACTCTTTCGATTCGTGTGGTTGGTACTAATAGCTTTGGTCGTAGAGGTTCTATATCCAGTGACATGGCAGTGCCAGACGGACGGTTGGCACGGCGGCTCTTTCGTCCGATTATACTTATGCGGTACCCTCACACTTCAGGCAGCCTTAATGCTGTTACTTGCTGCTTTGGCTCACCAGTCCGCCAGAGGGACCATTACAGATGTAGACATCAGAAAACTGGTATCACCATTACTATTAATCAAAGTGCTTCTGGTGTTGCCGGAAACAGCCCTCAATGTGATGGGTACTATGTGGATGTTCTGTGAAGTTATCGAGTGTTCAGTTGACGATAAATTCTCTAGCATAGTTATACAAAGTATAGTATTATTCAATTGGGTGCAACTAGGACTGACAGTGTTTGGGCTTTTCATGGTGTTCGATCCCTTGGGTTCAGTTAACTATGGAGATATGCAGGACACACCCAACCAAGTGCGCCATCATAGGAAAGTCACCGGTCTTTGGTCTCGGAGGTTCAGATGGGCGTTTTGTTGGCTGAAGAGAGACGAACATGGGAAGGAAGCATTTCAACAAGTTGCAGCACTGCTCAGCGCCTTGTTTCGATCCACGGACTTAGTACCATCTGACGTGGTAGCAGGATGTGTTCTGCTCAGAGTCAGACAGAAACGAGAAACTAGAGAAATGCGACGTATTCAAATGCTAAACGACGAAGAGCCTATTTATACGACGGACGTAAACAAGATATTCTCCGAAACCCCACCGTGGATGAATTTAGACGATGCCCTCTACTATCTCAGATTATCAATCGCTGCATATGGTTGGCCGTATGTTTTATACCGTCATTGCTTCACTGGCTTCTGCAAATTGGCCACCCACTTAACTTGTTGCTGTTGTCGTCCGAAAAATTCTATAGTAACGGACGACAACTGCTGTTTGTGTAACTTCGCTGGAGTCAAATACATGTCCAAATTACCTGCCGATGACATTATTTTCGCTTCCTTCAATAACAAAGTTTTTGAGTTGCCGTTCTGTGTGATAGCTGATCATGAACGCGAGTGTATAGTTGTGGCGGTGCGAGGGTCCATCTCACTCCGAGATATCTTCACGGATTTCACTGCTGGCTCCGAGAAATTTGAAGCTGACGGTTTACCAGAAAACACGGCAGCCCACAAAGGCATGGCAATGGGTGCGAACAAAATGTTGAAACGACTTTTACCGGTTCTCGACCGGACATTCCAGCAGTTTCCGCACTATGACCTAGTACTTACAGGTCACAGCTTAGGCGCTGGTGTAGCAGTATTAGTAGCTTTAAAATTACGACCGAGATACCCACATCTCAAAGTGTACGCGTTTTCAACGCCAGCGGGACTAATAAGTAGAGAAGCGGCGAGGTACACGGAGAGCTTTGTACTCACTGTGGGGGTCGGAGACGATTTGGTGATGAGGCTGAGCGTGCATAGTATAGAAAACCTTAGGACAAAAATCATTCAGACTATACACGCTACCAAACTACCTAAGTACCGAATAATGCTGAATGGCTTCGGTTACGCGTTGTTCGGTGTGCCCGCTCGCGACTTGGAGTCGACGTGGCGCCGCCCTGAGGACCTGGAGGCGACGCACTCGGACGACTCCGCTGACGCTCTCCTCGTGCCGAGCGTCTCCACCGAGGCTGCGTTAGTGTCGCGGGATATATTCGTGCGAAGATTCTCGTCAGCACGACTTTTCACTGCGGGGCGGATCTTGCACATCGCTCGACGTAAGCGCATGGGGATCGAAAAAAAAGTGCGCACGCAAGAGCCAACATATGAGATGCGCTGGGCCTGTCCCGAAGACTTCATGGAGTTACAAGTCATGCCTCGCATGCTATTGGACCATCTGCCCGAAAACGTTCATCGCACCATTCAAACAATAATCGAGGAGAGACATACTTACCGCGTCACGCACATAGTTTAG
Protein
MARMPALRLLGRKWLAASDDLVFPSIFELLFRFVWLVLIALVVEVLYPVTWQCQTDGWHGGSFVRLYLCGTLTLQAALMLLLAALAHQSARGTITDVDIRKLVSPLLLIKVLLVLPETALNVMGTMWMFCEVIECSVDDKFSSIVIQSIVLFNWVQLGLTVFGLFMVFDPLGSVNYGDMQDTPNQVRHHRKVTGLWSRRFRWAFCWLKRDEHGKEAFQQVAALLSALFRSTDLVPSDVVAGCVLLRVRQKRETREMRRIQMLNDEEPIYTTDVNKIFSETPPWMNLDDALYYLRLSIAAYGWPYVLYRHCFTGFCKLATHLTCCCCRPKNSIVTDDNCCLCNFAGVKYMSKLPADDIIFASFNNKVFELPFCVIADHERECIVVAVRGSISLRDIFTDFTAGSEKFEADGLPENTAAHKGMAMGANKMLKRLLPVLDRTFQQFPHYDLVLTGHSLGAGVAVLVALKLRPRYPHLKVYAFSTPAGLISREAARYTESFVLTVGVGDDLVMRLSVHSIENLRTKIIQTIHATKLPKYRIMLNGFGYALFGVPARDLESTWRRPEDLEATHSDDSADALLVPSVSTEAALVSRDIFVRRFSSARLFTAGRILHIARRKRMGIEKKVRTQEPTYEMRWACPEDFMELQVMPRMLLDHLPENVHRTIQTIIEERHTYRVTHIV
Summary
Uniprot
A0A2A4JWU9
A0A212FNM3
A0A194PE02
A0A194QMI0
A0A2H1V7L6
A0A3S2NEG6
+ More
H9JAL4 D6X1S2 A0A1Y1L0H4 A0A084WUK0 A0A1B0GLD8 A0A182YQ90 A0A1S4EW65 A0A2M4A5Z8 A0A2M4BFX2 A0A2M4BFC3 B0WU79 A0A1Q3FE43 A0A2M4BFH2 A0A182WD98 A0A1Q3FE28 W5JN64 A0A182X000 A0A182U4S3 A0A182MXB6 A0A182Q9H9 A0A182RAJ2 U4UFH7 A0A1B0D568 A0A182FTM0 E0V9X4 A0A182UTC2 A0A2C9GPH2 U5EY59 Q7PZZ9 Q17NZ4 A0A336MWK3 A0A336MJH1 A0A336L0H7 A0A182PGC5 A0A1J1J5R3 A0A2J7RRA8 K7IVM7 A0A232EQK5 A0A3L8DWK0 A0A026WPI9 E2A1B7 A0A336N1L9 A0A1B6CKF5 A0A195DXM6 A0A023F425 E2BZP4 A0A195BCP1 A0A151X6G7 A0A2H8TW89 F4X636 J9K315 A0A195FMG5 E9IFV3 A0A158NJQ6 A0A2S2QP66 A0A0C9RIU7 A0A154PHU5 A0A195D598 N6TA39 A0A0A9WM95 A0A1B6L3F1 A0A067RUY4 A0A310STX8 A0A1B6KSC0 A0A2J7RRA7 A0A224XLF7 A0A0L7QW72 A0A1J1IIS6 A0A1B6E9M6 A0A2A3E001 A0A088A4U1 A0A0A9ZB07 A0A146L6A4 A0A0K8TGF0 A0A2P8Z3K3 A0A1B6HS14 A0A1B6JFY7 A0A2R7X4T5 A0A3R7PXD3 E9GVS1
H9JAL4 D6X1S2 A0A1Y1L0H4 A0A084WUK0 A0A1B0GLD8 A0A182YQ90 A0A1S4EW65 A0A2M4A5Z8 A0A2M4BFX2 A0A2M4BFC3 B0WU79 A0A1Q3FE43 A0A2M4BFH2 A0A182WD98 A0A1Q3FE28 W5JN64 A0A182X000 A0A182U4S3 A0A182MXB6 A0A182Q9H9 A0A182RAJ2 U4UFH7 A0A1B0D568 A0A182FTM0 E0V9X4 A0A182UTC2 A0A2C9GPH2 U5EY59 Q7PZZ9 Q17NZ4 A0A336MWK3 A0A336MJH1 A0A336L0H7 A0A182PGC5 A0A1J1J5R3 A0A2J7RRA8 K7IVM7 A0A232EQK5 A0A3L8DWK0 A0A026WPI9 E2A1B7 A0A336N1L9 A0A1B6CKF5 A0A195DXM6 A0A023F425 E2BZP4 A0A195BCP1 A0A151X6G7 A0A2H8TW89 F4X636 J9K315 A0A195FMG5 E9IFV3 A0A158NJQ6 A0A2S2QP66 A0A0C9RIU7 A0A154PHU5 A0A195D598 N6TA39 A0A0A9WM95 A0A1B6L3F1 A0A067RUY4 A0A310STX8 A0A1B6KSC0 A0A2J7RRA7 A0A224XLF7 A0A0L7QW72 A0A1J1IIS6 A0A1B6E9M6 A0A2A3E001 A0A088A4U1 A0A0A9ZB07 A0A146L6A4 A0A0K8TGF0 A0A2P8Z3K3 A0A1B6HS14 A0A1B6JFY7 A0A2R7X4T5 A0A3R7PXD3 E9GVS1
Pubmed
EMBL
NWSH01000455
PCG76289.1
AGBW02005235
OWR55327.1
KQ459606
KPI91566.1
+ More
KQ461198 KPJ06160.1 ODYU01001079 SOQ36811.1 RSAL01000073 RVE48986.1 BABH01008753 KQ971371 EFA10154.1 GEZM01068218 GEZM01068217 JAV67084.1 ATLV01027094 KE525423 KFB53894.1 AJWK01034779 AJWK01034780 GGFK01002915 MBW36236.1 GGFJ01002815 MBW51956.1 GGFJ01002608 MBW51749.1 DS232102 EDS34851.1 GFDL01009221 JAV25824.1 GGFJ01002641 MBW51782.1 GFDL01009225 JAV25820.1 ADMH02001047 ETN64330.1 AXCN02001682 KB632092 ERL88670.1 AJVK01003331 AJVK01003332 DS235000 EEB10180.1 APCN01003270 GANO01000578 JAB59293.1 AAAB01008986 EAA00159.4 CH477195 EAT48404.1 UFQT01003133 SSX34626.1 UFQS01001411 UFQT01001411 SSX10754.1 SSX30436.1 SSX10755.1 SSX30437.1 CVRI01000073 CRL07735.1 NEVH01000610 PNF43369.1 NNAY01002745 OXU20644.1 QOIP01000003 RLU24821.1 KK107144 EZA57566.1 GL435746 EFN72777.1 SSX34627.1 GEDC01025992 GEDC01023382 JAS11306.1 JAS13916.1 KQ980155 KYN17467.1 GBBI01002983 JAC15729.1 GL451645 EFN78859.1 KQ976526 KYM81960.1 KQ982482 KYQ55909.1 GFXV01005623 MBW17428.1 GL888763 EGI58098.1 ABLF02036577 KQ981424 KYN41860.1 GL762903 EFZ20552.1 ADTU01018184 ADTU01018185 GGMS01010363 MBY79566.1 GBYB01013162 JAG82929.1 KQ434900 KZC11064.1 KQ976818 KYN08075.1 APGK01038131 APGK01038132 APGK01038133 KB740953 ENN77119.1 GBHO01034047 GBHO01034046 GBHO01034044 GBHO01034042 GDHC01006198 GDHC01003484 GDHC01001233 JAG09557.1 JAG09558.1 JAG09560.1 JAG09562.1 JAQ12431.1 JAQ15145.1 JAQ17396.1 GEBQ01021734 JAT18243.1 KK852453 KDR23659.1 KQ759891 OAD62108.1 GEBQ01025638 JAT14339.1 PNF43368.1 GFTR01007567 JAW08859.1 KQ414716 KOC62809.1 CVRI01000054 CRL00123.1 GEDC01003538 GEDC01002660 JAS33760.1 JAS34638.1 KZ288522 PBC25090.1 GBHO01022391 GBHO01001142 GBHO01001141 GBHO01001137 JAG21213.1 JAG42462.1 JAG42463.1 JAG42467.1 GDHC01014651 JAQ03978.1 GBRD01001345 JAG64476.1 PYGN01000212 PSN51081.1 GECU01030266 JAS77440.1 GECU01009591 JAS98115.1 KK857087 PTY26804.1 QCYY01001152 ROT80111.1 GL732568 EFX76477.1
KQ461198 KPJ06160.1 ODYU01001079 SOQ36811.1 RSAL01000073 RVE48986.1 BABH01008753 KQ971371 EFA10154.1 GEZM01068218 GEZM01068217 JAV67084.1 ATLV01027094 KE525423 KFB53894.1 AJWK01034779 AJWK01034780 GGFK01002915 MBW36236.1 GGFJ01002815 MBW51956.1 GGFJ01002608 MBW51749.1 DS232102 EDS34851.1 GFDL01009221 JAV25824.1 GGFJ01002641 MBW51782.1 GFDL01009225 JAV25820.1 ADMH02001047 ETN64330.1 AXCN02001682 KB632092 ERL88670.1 AJVK01003331 AJVK01003332 DS235000 EEB10180.1 APCN01003270 GANO01000578 JAB59293.1 AAAB01008986 EAA00159.4 CH477195 EAT48404.1 UFQT01003133 SSX34626.1 UFQS01001411 UFQT01001411 SSX10754.1 SSX30436.1 SSX10755.1 SSX30437.1 CVRI01000073 CRL07735.1 NEVH01000610 PNF43369.1 NNAY01002745 OXU20644.1 QOIP01000003 RLU24821.1 KK107144 EZA57566.1 GL435746 EFN72777.1 SSX34627.1 GEDC01025992 GEDC01023382 JAS11306.1 JAS13916.1 KQ980155 KYN17467.1 GBBI01002983 JAC15729.1 GL451645 EFN78859.1 KQ976526 KYM81960.1 KQ982482 KYQ55909.1 GFXV01005623 MBW17428.1 GL888763 EGI58098.1 ABLF02036577 KQ981424 KYN41860.1 GL762903 EFZ20552.1 ADTU01018184 ADTU01018185 GGMS01010363 MBY79566.1 GBYB01013162 JAG82929.1 KQ434900 KZC11064.1 KQ976818 KYN08075.1 APGK01038131 APGK01038132 APGK01038133 KB740953 ENN77119.1 GBHO01034047 GBHO01034046 GBHO01034044 GBHO01034042 GDHC01006198 GDHC01003484 GDHC01001233 JAG09557.1 JAG09558.1 JAG09560.1 JAG09562.1 JAQ12431.1 JAQ15145.1 JAQ17396.1 GEBQ01021734 JAT18243.1 KK852453 KDR23659.1 KQ759891 OAD62108.1 GEBQ01025638 JAT14339.1 PNF43368.1 GFTR01007567 JAW08859.1 KQ414716 KOC62809.1 CVRI01000054 CRL00123.1 GEDC01003538 GEDC01002660 JAS33760.1 JAS34638.1 KZ288522 PBC25090.1 GBHO01022391 GBHO01001142 GBHO01001141 GBHO01001137 JAG21213.1 JAG42462.1 JAG42463.1 JAG42467.1 GDHC01014651 JAQ03978.1 GBRD01001345 JAG64476.1 PYGN01000212 PSN51081.1 GECU01030266 JAS77440.1 GECU01009591 JAS98115.1 KK857087 PTY26804.1 QCYY01001152 ROT80111.1 GL732568 EFX76477.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000283053
UP000005204
+ More
UP000007266 UP000030765 UP000092461 UP000076408 UP000002320 UP000075920 UP000000673 UP000076407 UP000075902 UP000075884 UP000075886 UP000075900 UP000030742 UP000092462 UP000069272 UP000009046 UP000075903 UP000075840 UP000007062 UP000008820 UP000075885 UP000183832 UP000235965 UP000002358 UP000215335 UP000279307 UP000053097 UP000000311 UP000078492 UP000008237 UP000078540 UP000075809 UP000007755 UP000007819 UP000078541 UP000005205 UP000076502 UP000078542 UP000019118 UP000027135 UP000053825 UP000242457 UP000005203 UP000245037 UP000283509 UP000000305
UP000007266 UP000030765 UP000092461 UP000076408 UP000002320 UP000075920 UP000000673 UP000076407 UP000075902 UP000075884 UP000075886 UP000075900 UP000030742 UP000092462 UP000069272 UP000009046 UP000075903 UP000075840 UP000007062 UP000008820 UP000075885 UP000183832 UP000235965 UP000002358 UP000215335 UP000279307 UP000053097 UP000000311 UP000078492 UP000008237 UP000078540 UP000075809 UP000007755 UP000007819 UP000078541 UP000005205 UP000076502 UP000078542 UP000019118 UP000027135 UP000053825 UP000242457 UP000005203 UP000245037 UP000283509 UP000000305
PRIDE
Pfam
PF01764 Lipase_3
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2A4JWU9
A0A212FNM3
A0A194PE02
A0A194QMI0
A0A2H1V7L6
A0A3S2NEG6
+ More
H9JAL4 D6X1S2 A0A1Y1L0H4 A0A084WUK0 A0A1B0GLD8 A0A182YQ90 A0A1S4EW65 A0A2M4A5Z8 A0A2M4BFX2 A0A2M4BFC3 B0WU79 A0A1Q3FE43 A0A2M4BFH2 A0A182WD98 A0A1Q3FE28 W5JN64 A0A182X000 A0A182U4S3 A0A182MXB6 A0A182Q9H9 A0A182RAJ2 U4UFH7 A0A1B0D568 A0A182FTM0 E0V9X4 A0A182UTC2 A0A2C9GPH2 U5EY59 Q7PZZ9 Q17NZ4 A0A336MWK3 A0A336MJH1 A0A336L0H7 A0A182PGC5 A0A1J1J5R3 A0A2J7RRA8 K7IVM7 A0A232EQK5 A0A3L8DWK0 A0A026WPI9 E2A1B7 A0A336N1L9 A0A1B6CKF5 A0A195DXM6 A0A023F425 E2BZP4 A0A195BCP1 A0A151X6G7 A0A2H8TW89 F4X636 J9K315 A0A195FMG5 E9IFV3 A0A158NJQ6 A0A2S2QP66 A0A0C9RIU7 A0A154PHU5 A0A195D598 N6TA39 A0A0A9WM95 A0A1B6L3F1 A0A067RUY4 A0A310STX8 A0A1B6KSC0 A0A2J7RRA7 A0A224XLF7 A0A0L7QW72 A0A1J1IIS6 A0A1B6E9M6 A0A2A3E001 A0A088A4U1 A0A0A9ZB07 A0A146L6A4 A0A0K8TGF0 A0A2P8Z3K3 A0A1B6HS14 A0A1B6JFY7 A0A2R7X4T5 A0A3R7PXD3 E9GVS1
H9JAL4 D6X1S2 A0A1Y1L0H4 A0A084WUK0 A0A1B0GLD8 A0A182YQ90 A0A1S4EW65 A0A2M4A5Z8 A0A2M4BFX2 A0A2M4BFC3 B0WU79 A0A1Q3FE43 A0A2M4BFH2 A0A182WD98 A0A1Q3FE28 W5JN64 A0A182X000 A0A182U4S3 A0A182MXB6 A0A182Q9H9 A0A182RAJ2 U4UFH7 A0A1B0D568 A0A182FTM0 E0V9X4 A0A182UTC2 A0A2C9GPH2 U5EY59 Q7PZZ9 Q17NZ4 A0A336MWK3 A0A336MJH1 A0A336L0H7 A0A182PGC5 A0A1J1J5R3 A0A2J7RRA8 K7IVM7 A0A232EQK5 A0A3L8DWK0 A0A026WPI9 E2A1B7 A0A336N1L9 A0A1B6CKF5 A0A195DXM6 A0A023F425 E2BZP4 A0A195BCP1 A0A151X6G7 A0A2H8TW89 F4X636 J9K315 A0A195FMG5 E9IFV3 A0A158NJQ6 A0A2S2QP66 A0A0C9RIU7 A0A154PHU5 A0A195D598 N6TA39 A0A0A9WM95 A0A1B6L3F1 A0A067RUY4 A0A310STX8 A0A1B6KSC0 A0A2J7RRA7 A0A224XLF7 A0A0L7QW72 A0A1J1IIS6 A0A1B6E9M6 A0A2A3E001 A0A088A4U1 A0A0A9ZB07 A0A146L6A4 A0A0K8TGF0 A0A2P8Z3K3 A0A1B6HS14 A0A1B6JFY7 A0A2R7X4T5 A0A3R7PXD3 E9GVS1
PDB
5CH8
E-value=1.62479e-09,
Score=152
Ontologies
GO
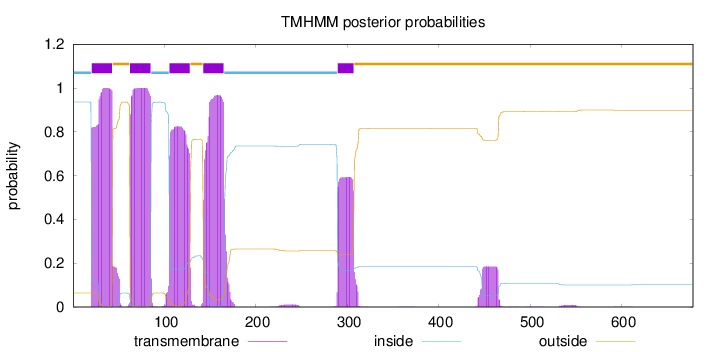
Topology
Length:
678
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
101.75976
Exp number, first 60 AAs:
22.80813
Total prob of N-in:
0.93598
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 62
TMhelix
63 - 85
inside
86 - 105
TMhelix
106 - 128
outside
129 - 142
TMhelix
143 - 165
inside
166 - 289
TMhelix
290 - 307
outside
308 - 678
Population Genetic Test Statistics
Pi
219.251652
Theta
169.054193
Tajima's D
0.90766
CLR
0.201673
CSRT
0.64206789660517
Interpretation
Uncertain
