Gene
KWMTBOMO05622
Pre Gene Modal
BGIBMGA006561
Annotation
PREDICTED:_solute_carrier_family_22_member_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.894
Sequence
CDS
ATGGCATTTAACAATATAATTCTGGCCCTCACGATTATTCCCTATACTTGTGAGATACCAGACGCTCCTAAGAATGTGAACAGAGAAGATTGGAAGGAAATGTGGATTCCGAAGAATCAGTTAGGCGGCAATATGACAATATTCAGCAATTGCTTAATTTATGACGAAAATAACAACATCACAGATTGCAAACGTTACCGATACGATAATACTTGGTACAAAAGTACGATTCCTTCAGAGAACAATTGGGTGTGCGAAAAAGAATATTATGTAATAAACATTTTATCACACAACAAGATTTCTGAAACTGTTGGATCACTTATCTTTGGTTGGTTTGGTGATGCGTATGGACGACGGTTAACTATAGGATGTTTAATAGCCGCTTTCCCAGCGACTTCTGCCTTACAATGCACATCTTCGATTTCAATGGAGATTTCATCTCCTAAAAGAAGATCGAGTATCGCTAAATTACGATTTATTGCTTCAAGTGTTGGTTTGTGCCTGATGCCCCTATTATATTGGTGGATAAGAGAATGGAAATTATTTATGATTACGACGACAGTCACGCAATTGCCATATTTACTGTGGTCGTGGAAGATGATCGAATCTCCGCAATGGTTGTGGATAAATCAAAGAAACAAAGAGTGTGAGAACATTTTAGAGCAAATAGCGAAAGACAATCGCACTGCGTTGAGCAGCCAAACAAAACAATTGATCTCAGCATCGCACGAAGAACACACCGAAAAACAATTTGGATTCCTTCTTCTTTTTTCCAACAAGGAATTAGCCCTTGTTACGATTTTACAGCTTTTTATATGGGTATCAGTAACATTAAGTTACACCGTCAGCTTTTTAAGTTCAGGAGAAAGCTCACAAACAAATCCTTTTTTAGATTTTGCTTTTCAGTCACTTGCTGAAATACCAGGGCACTTTTCAGGAGCTTGGTTAGCCGATAATATAGGTCGTCGGTATACCTGTGTAGTTTCATTTGGTGTGTCGTCTGTTGCATGGGTCATAATTGGTTTCAAAGATTTTGTCATGTGGCCAGATGTCGAATGGATTTTGATATTCGTACAGGTTTTAAATAGATACTCTATAACTATATCTTACTACATTATAAATCTACTTAATATGGAAATTTATCCTACTGCCATGCGTCAGTCTGGAATGGCTTTGGGCAATTTGATTTCTGGAATAGCGGCAGCGATTGCTCCCTATGTACTTTTATTGGGACATAAACACGGATCCTTTTGGTCTTCAGCCATCCTAAGCCTGTTCTCAATAATAGGATTGATGTCAGGCTTAGGCCTACCAGAGACAGTAGCTCGCAGGAGACATCCAATTTGA
Protein
MAFNNIILALTIIPYTCEIPDAPKNVNREDWKEMWIPKNQLGGNMTIFSNCLIYDENNNITDCKRYRYDNTWYKSTIPSENNWVCEKEYYVINILSHNKISETVGSLIFGWFGDAYGRRLTIGCLIAAFPATSALQCTSSISMEISSPKRRSSIAKLRFIASSVGLCLMPLLYWWIREWKLFMITTTVTQLPYLLWSWKMIESPQWLWINQRNKECENILEQIAKDNRTALSSQTKQLISASHEEHTEKQFGFLLLFSNKELALVTILQLFIWVSVTLSYTVSFLSSGESSQTNPFLDFAFQSLAEIPGHFSGAWLADNIGRRYTCVVSFGVSSVAWVIIGFKDFVMWPDVEWILIFVQVLNRYSITISYYIINLLNMEIYPTAMRQSGMALGNLISGIAAAIAPYVLLLGHKHGSFWSSAILSLFSIIGLMSGLGLPETVARRRHPI
Summary
Uniprot
A0A2H1WJY9
A0A3S2TL02
A0A2A4JW86
A0A0M4ERY9
B4PLL6
B3P7M1
+ More
B4QZY1 B4HE52 Q9VD40 B4JRN1 A0A1W4W3D5 B4M0E5 B4NHS7 A0A0K8U2L8 A0A0K8W4N1 A0A034VFA1 B4K6N3 A0A1A9YCB9 A0A1B0FQN9 A0A3B0JG24 A0A0A1WKV3 Q299K7 T1P920 B3LVM6 A0A0L0CJ20 A0A1I8PE87 A0A1I8NHI3 W8B981 A0A336L3U2 A0A336MX72 A0A336L090 A0A2J7PP99 D6WD21 A0A2J7RAH2 A0A1J1HR55 A0A1W4WQ60 A0A0Q9XAL6 A0A067QUZ6 A0A1L8DE32 A0A182QNX3 A0A336LT19 A0A1A9ZQ56 A0A1S4F435 A0A1A9WDS2 F5HL30 F5HL29 A0A182XFK7 A0A182Y744 A0A182GEV7 A0A182UTZ0 A0A182F777 A0A084VCH7 A0A2J7RAG8 A0A2M4BHW1 A0A2M4BIK2 Q17G50 B0XD15 A0A2M4BHY4 A0A2M4BHW3 A0A1Q3FRL9 A0A1Q3FRP7 W5JDH3 A0A2J7PPD9 A0A182U8Z6 A0A182RIC5 A0A182GFZ2 A0A182I7Y9 A0A182W0H0 A0A182QL49 B0WY17 Q17G56 A0A182N5L2 A0A2M4ALR2 A0A182J0N1 A0A182VL36 A0A182Y747 A0A084VCW6 E0VJD0 A0A2J7PPC2 A0A182INI2 A0A182XFK3 A0A182I7Z3 A0A182L7K6 A0A182RIC1 Q7PQK9 A0A182N5L6 A0A182U5R3 A0A182F782 A0A2M4BKQ0 A0A2M4APY9 A0A182JSI3 Q17G55 A0A182W0H4 A0A182PNN3 A0A182M191 A0A0K8TYT1 A0A182PNN8 A0A182JWC4 A0A1I8MU15
B4QZY1 B4HE52 Q9VD40 B4JRN1 A0A1W4W3D5 B4M0E5 B4NHS7 A0A0K8U2L8 A0A0K8W4N1 A0A034VFA1 B4K6N3 A0A1A9YCB9 A0A1B0FQN9 A0A3B0JG24 A0A0A1WKV3 Q299K7 T1P920 B3LVM6 A0A0L0CJ20 A0A1I8PE87 A0A1I8NHI3 W8B981 A0A336L3U2 A0A336MX72 A0A336L090 A0A2J7PP99 D6WD21 A0A2J7RAH2 A0A1J1HR55 A0A1W4WQ60 A0A0Q9XAL6 A0A067QUZ6 A0A1L8DE32 A0A182QNX3 A0A336LT19 A0A1A9ZQ56 A0A1S4F435 A0A1A9WDS2 F5HL30 F5HL29 A0A182XFK7 A0A182Y744 A0A182GEV7 A0A182UTZ0 A0A182F777 A0A084VCH7 A0A2J7RAG8 A0A2M4BHW1 A0A2M4BIK2 Q17G50 B0XD15 A0A2M4BHY4 A0A2M4BHW3 A0A1Q3FRL9 A0A1Q3FRP7 W5JDH3 A0A2J7PPD9 A0A182U8Z6 A0A182RIC5 A0A182GFZ2 A0A182I7Y9 A0A182W0H0 A0A182QL49 B0WY17 Q17G56 A0A182N5L2 A0A2M4ALR2 A0A182J0N1 A0A182VL36 A0A182Y747 A0A084VCW6 E0VJD0 A0A2J7PPC2 A0A182INI2 A0A182XFK3 A0A182I7Z3 A0A182L7K6 A0A182RIC1 Q7PQK9 A0A182N5L6 A0A182U5R3 A0A182F782 A0A2M4BKQ0 A0A2M4APY9 A0A182JSI3 Q17G55 A0A182W0H4 A0A182PNN3 A0A182M191 A0A0K8TYT1 A0A182PNN8 A0A182JWC4 A0A1I8MU15
Pubmed
17994087
17550304
10731132
12537568
12537572
12537573
+ More
12537574 16110336 17569856 17569867 26109357 26109356 18057021 25348373 25830018 15632085 26108605 25315136 24495485 18362917 19820115 24845553 17510324 12364791 14747013 17210077 25244985 26483478 24438588 20920257 23761445 20566863 20966253
12537574 16110336 17569856 17569867 26109357 26109356 18057021 25348373 25830018 15632085 26108605 25315136 24495485 18362917 19820115 24845553 17510324 12364791 14747013 17210077 25244985 26483478 24438588 20920257 23761445 20566863 20966253
EMBL
ODYU01009143
SOQ53358.1
RSAL01000073
RVE48995.1
NWSH01000455
PCG76297.1
+ More
CP012526 ALC47410.1 CM000160 EDW97965.1 KRK04041.1 CH954182 EDV54182.1 CM000364 EDX13887.1 CH480815 EDW43149.1 AE014297 BT009957 AAF55963.1 AAQ22426.1 CH916373 EDV94421.1 CH940650 EDW67307.1 KRF83233.1 CH964272 EDW84687.1 GDHF01031719 JAI20595.1 GDHF01019328 GDHF01014238 GDHF01006146 JAI32986.1 JAI38076.1 JAI46168.1 GAKP01018745 GAKP01018744 GAKP01018743 JAC40208.1 CH933806 EDW16333.1 CCAG010022561 OUUW01000005 SPP81344.1 GBXI01016221 GBXI01015142 JAC98070.1 JAC99149.1 CM000070 EAL27696.2 KA645194 AFP59823.1 CH902617 EDV42596.1 JRES01000328 KNC32251.1 GAMC01012947 JAB93608.1 UFQS01001486 UFQT01001486 SSX11199.1 SSX30769.1 UFQT01002357 SSX33283.1 SSX11200.1 SSX30770.1 NEVH01023278 PNF18174.1 KQ971311 EEZ98813.2 NEVH01006570 PNF37820.1 CVRI01000010 CRK88958.1 KRG02052.1 KK853234 KDR09584.1 GFDF01009484 JAV04600.1 AXCN02001160 UFQS01005318 UFQT01005318 SSX16788.1 SSX35974.1 AAAB01008880 EGK96989.1 EAL40645.4 EGK96990.1 EGK96991.1 JXUM01058636 JXUM01058637 JXUM01058638 KQ562011 KXJ76913.1 ATLV01010759 KE524612 KFB35671.1 PNF37824.1 GGFJ01003509 MBW52650.1 GGFJ01003507 MBW52648.1 CH477266 EAT45535.1 DS232734 EDS45250.1 GGFJ01003506 MBW52647.1 GGFJ01003508 MBW52649.1 GFDL01004796 JAV30249.1 GFDL01004942 JAV30103.1 ADMH02001534 ETN62151.1 PNF18176.1 JXUM01060980 KQ562129 KXJ76620.1 APCN01002589 AXCN02001161 DS232179 EDS36827.1 EAT45528.1 GGFK01008399 MBW41720.1 ATLV01010943 KE524624 KFB35810.1 DS235222 EEB13486.1 PNF18178.1 APCN01002591 EAA08673.4 EGK96988.1 GGFJ01004506 MBW53647.1 GGFK01009357 MBW42678.1 EAT45529.1 AXCM01002248 GDHF01032692 GDHF01012127 JAI19622.1 JAI40187.1
CP012526 ALC47410.1 CM000160 EDW97965.1 KRK04041.1 CH954182 EDV54182.1 CM000364 EDX13887.1 CH480815 EDW43149.1 AE014297 BT009957 AAF55963.1 AAQ22426.1 CH916373 EDV94421.1 CH940650 EDW67307.1 KRF83233.1 CH964272 EDW84687.1 GDHF01031719 JAI20595.1 GDHF01019328 GDHF01014238 GDHF01006146 JAI32986.1 JAI38076.1 JAI46168.1 GAKP01018745 GAKP01018744 GAKP01018743 JAC40208.1 CH933806 EDW16333.1 CCAG010022561 OUUW01000005 SPP81344.1 GBXI01016221 GBXI01015142 JAC98070.1 JAC99149.1 CM000070 EAL27696.2 KA645194 AFP59823.1 CH902617 EDV42596.1 JRES01000328 KNC32251.1 GAMC01012947 JAB93608.1 UFQS01001486 UFQT01001486 SSX11199.1 SSX30769.1 UFQT01002357 SSX33283.1 SSX11200.1 SSX30770.1 NEVH01023278 PNF18174.1 KQ971311 EEZ98813.2 NEVH01006570 PNF37820.1 CVRI01000010 CRK88958.1 KRG02052.1 KK853234 KDR09584.1 GFDF01009484 JAV04600.1 AXCN02001160 UFQS01005318 UFQT01005318 SSX16788.1 SSX35974.1 AAAB01008880 EGK96989.1 EAL40645.4 EGK96990.1 EGK96991.1 JXUM01058636 JXUM01058637 JXUM01058638 KQ562011 KXJ76913.1 ATLV01010759 KE524612 KFB35671.1 PNF37824.1 GGFJ01003509 MBW52650.1 GGFJ01003507 MBW52648.1 CH477266 EAT45535.1 DS232734 EDS45250.1 GGFJ01003506 MBW52647.1 GGFJ01003508 MBW52649.1 GFDL01004796 JAV30249.1 GFDL01004942 JAV30103.1 ADMH02001534 ETN62151.1 PNF18176.1 JXUM01060980 KQ562129 KXJ76620.1 APCN01002589 AXCN02001161 DS232179 EDS36827.1 EAT45528.1 GGFK01008399 MBW41720.1 ATLV01010943 KE524624 KFB35810.1 DS235222 EEB13486.1 PNF18178.1 APCN01002591 EAA08673.4 EGK96988.1 GGFJ01004506 MBW53647.1 GGFK01009357 MBW42678.1 EAT45529.1 AXCM01002248 GDHF01032692 GDHF01012127 JAI19622.1 JAI40187.1
Proteomes
UP000283053
UP000218220
UP000092553
UP000002282
UP000008711
UP000000304
+ More
UP000001292 UP000000803 UP000001070 UP000192221 UP000008792 UP000007798 UP000009192 UP000092443 UP000092444 UP000268350 UP000001819 UP000007801 UP000037069 UP000095300 UP000095301 UP000235965 UP000007266 UP000183832 UP000192223 UP000027135 UP000075886 UP000092445 UP000091820 UP000007062 UP000076407 UP000076408 UP000069940 UP000249989 UP000075903 UP000069272 UP000030765 UP000008820 UP000002320 UP000000673 UP000075902 UP000075900 UP000075840 UP000075920 UP000075884 UP000075880 UP000009046 UP000075882 UP000075881 UP000075885 UP000075883
UP000001292 UP000000803 UP000001070 UP000192221 UP000008792 UP000007798 UP000009192 UP000092443 UP000092444 UP000268350 UP000001819 UP000007801 UP000037069 UP000095300 UP000095301 UP000235965 UP000007266 UP000183832 UP000192223 UP000027135 UP000075886 UP000092445 UP000091820 UP000007062 UP000076407 UP000076408 UP000069940 UP000249989 UP000075903 UP000069272 UP000030765 UP000008820 UP000002320 UP000000673 UP000075902 UP000075900 UP000075840 UP000075920 UP000075884 UP000075880 UP000009046 UP000075882 UP000075881 UP000075885 UP000075883
PRIDE
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1WJY9
A0A3S2TL02
A0A2A4JW86
A0A0M4ERY9
B4PLL6
B3P7M1
+ More
B4QZY1 B4HE52 Q9VD40 B4JRN1 A0A1W4W3D5 B4M0E5 B4NHS7 A0A0K8U2L8 A0A0K8W4N1 A0A034VFA1 B4K6N3 A0A1A9YCB9 A0A1B0FQN9 A0A3B0JG24 A0A0A1WKV3 Q299K7 T1P920 B3LVM6 A0A0L0CJ20 A0A1I8PE87 A0A1I8NHI3 W8B981 A0A336L3U2 A0A336MX72 A0A336L090 A0A2J7PP99 D6WD21 A0A2J7RAH2 A0A1J1HR55 A0A1W4WQ60 A0A0Q9XAL6 A0A067QUZ6 A0A1L8DE32 A0A182QNX3 A0A336LT19 A0A1A9ZQ56 A0A1S4F435 A0A1A9WDS2 F5HL30 F5HL29 A0A182XFK7 A0A182Y744 A0A182GEV7 A0A182UTZ0 A0A182F777 A0A084VCH7 A0A2J7RAG8 A0A2M4BHW1 A0A2M4BIK2 Q17G50 B0XD15 A0A2M4BHY4 A0A2M4BHW3 A0A1Q3FRL9 A0A1Q3FRP7 W5JDH3 A0A2J7PPD9 A0A182U8Z6 A0A182RIC5 A0A182GFZ2 A0A182I7Y9 A0A182W0H0 A0A182QL49 B0WY17 Q17G56 A0A182N5L2 A0A2M4ALR2 A0A182J0N1 A0A182VL36 A0A182Y747 A0A084VCW6 E0VJD0 A0A2J7PPC2 A0A182INI2 A0A182XFK3 A0A182I7Z3 A0A182L7K6 A0A182RIC1 Q7PQK9 A0A182N5L6 A0A182U5R3 A0A182F782 A0A2M4BKQ0 A0A2M4APY9 A0A182JSI3 Q17G55 A0A182W0H4 A0A182PNN3 A0A182M191 A0A0K8TYT1 A0A182PNN8 A0A182JWC4 A0A1I8MU15
B4QZY1 B4HE52 Q9VD40 B4JRN1 A0A1W4W3D5 B4M0E5 B4NHS7 A0A0K8U2L8 A0A0K8W4N1 A0A034VFA1 B4K6N3 A0A1A9YCB9 A0A1B0FQN9 A0A3B0JG24 A0A0A1WKV3 Q299K7 T1P920 B3LVM6 A0A0L0CJ20 A0A1I8PE87 A0A1I8NHI3 W8B981 A0A336L3U2 A0A336MX72 A0A336L090 A0A2J7PP99 D6WD21 A0A2J7RAH2 A0A1J1HR55 A0A1W4WQ60 A0A0Q9XAL6 A0A067QUZ6 A0A1L8DE32 A0A182QNX3 A0A336LT19 A0A1A9ZQ56 A0A1S4F435 A0A1A9WDS2 F5HL30 F5HL29 A0A182XFK7 A0A182Y744 A0A182GEV7 A0A182UTZ0 A0A182F777 A0A084VCH7 A0A2J7RAG8 A0A2M4BHW1 A0A2M4BIK2 Q17G50 B0XD15 A0A2M4BHY4 A0A2M4BHW3 A0A1Q3FRL9 A0A1Q3FRP7 W5JDH3 A0A2J7PPD9 A0A182U8Z6 A0A182RIC5 A0A182GFZ2 A0A182I7Y9 A0A182W0H0 A0A182QL49 B0WY17 Q17G56 A0A182N5L2 A0A2M4ALR2 A0A182J0N1 A0A182VL36 A0A182Y747 A0A084VCW6 E0VJD0 A0A2J7PPC2 A0A182INI2 A0A182XFK3 A0A182I7Z3 A0A182L7K6 A0A182RIC1 Q7PQK9 A0A182N5L6 A0A182U5R3 A0A182F782 A0A2M4BKQ0 A0A2M4APY9 A0A182JSI3 Q17G55 A0A182W0H4 A0A182PNN3 A0A182M191 A0A0K8TYT1 A0A182PNN8 A0A182JWC4 A0A1I8MU15
PDB
6N3I
E-value=0.000333368,
Score=105
Ontologies
GO
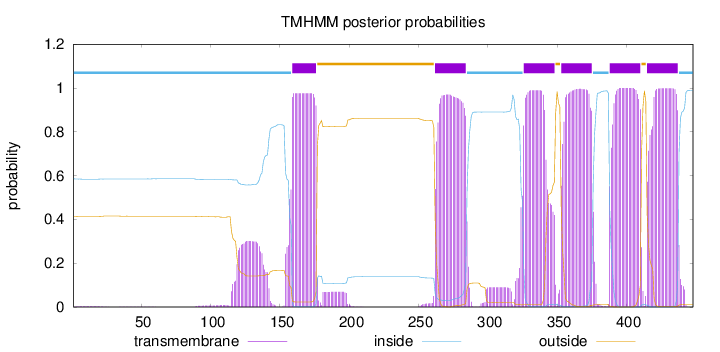
Topology
Length:
448
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
139.05012
Exp number, first 60 AAs:
0.07112
Total prob of N-in:
0.58620
inside
1 - 158
TMhelix
159 - 176
outside
177 - 261
TMhelix
262 - 284
inside
285 - 325
TMhelix
326 - 348
outside
349 - 352
TMhelix
353 - 375
inside
376 - 387
TMhelix
388 - 410
outside
411 - 414
TMhelix
415 - 437
inside
438 - 448
Population Genetic Test Statistics
Pi
209.844799
Theta
180.084209
Tajima's D
0.729724
CLR
0.020759
CSRT
0.579171041447928
Interpretation
Uncertain
