Gene
KWMTBOMO05621
Pre Gene Modal
BGIBMGA006562
Annotation
PREDICTED:_BTB/POZ_domain-containing_adapter_for_CUL3-mediated_RhoA_degradation_protein_3_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.363
Sequence
CDS
ATGTTATTTCTTCGTTTTGAAATAATGTCGGGAGATCACAAAACGTTGTTAAAAGGCAGTCCGTCACAATATGTGAAGTTAAATGTGGGCGGCACATTGTTCTACACTACTATCGGCACGCTTACGAAAAGCGATAACATGTTGCGAACCATGTTTAGTGGGAGGATGGAGGTTCTCACAGATTCGGAAGGGTGGATTTTGATTGATCGTTGTGGGAAGCATTTCGGTACTATTTTGAATTATCTTCGTGACGGCACAGTAGCGCTACCCGACAGTTATAAAGAGATCATGGAGTTATTGGCGGAGGCTAAATATTTCCTGATTGAAGAGTTAACTGAATCTTGCCTACAGGCCTTAGCCAAAAAAGAACGCGAGGCAGAGCCTATTTGCAGAGTTCCTTTAATTACTTCACAAAAGGAAGAACAACTTTTAATTATGTCCACTTCCAAGCCTGTGGTGAAACTGTTGATTAACAGACACAATAACAAGTATTCTTACACGAGCACTTCTGATGATAACTTACTAAAGAACATTGAACTCTTTGACAAACTTTCGCTGCGATTCAGCGGGAGAGTGCTGTTTATTAAAGATGTTATTGGTTCTAATGAGATTTGTTGTTGGTCTTTCTTTGGACATGGAAAAAAATGTGCTGAAGTCTGCTGCACTTCAATTGTGTATGCTACAGATAAAAAGCATACTAAGGTTGAGTTTCCGGAGGCCAGGATATATGAGGAAACACTGGGAATTTTACTTTATGAAAGTCGTAATGGCCCTGATCAAGATTTAATTCAAGCTACTTCATCACGAGGTGCTGTTGGGGGATTGTCTTGCACTAGTGATGAGGAGGAAGAGCGATCTGGCCTTGCCCGACTTAGATCAAATAAGCAGAATAATCAATCTTAG
Protein
MLFLRFEIMSGDHKTLLKGSPSQYVKLNVGGTLFYTTIGTLTKSDNMLRTMFSGRMEVLTDSEGWILIDRCGKHFGTILNYLRDGTVALPDSYKEIMELLAEAKYFLIEELTESCLQALAKKEREAEPICRVPLITSQKEEQLLIMSTSKPVVKLLINRHNNKYSYTSTSDDNLLKNIELFDKLSLRFSGRVLFIKDVIGSNEICCWSFFGHGKKCAEVCCTSIVYATDKKHTKVEFPEARIYEETLGILLYESRNGPDQDLIQATSSRGAVGGLSCTSDEEEERSGLARLRSNKQNNQS
Summary
Uniprot
H9JAL7
A0A2A4JX72
A0A2H1WJY7
A0A1E1WP15
A0A212FNK9
A0A3S2M1R6
+ More
A0A194QMI5 A0A194PE83 S4PFJ4 A0A0L7KWP3 A0A067R7M6 A0A1B6FLY3 A0A1B6J6G6 A0A2J7QTS5 A0A0N7Z9B0 R4G4Q0 A0A224XGY7 A0A023F9T2 E2B9K5 A0A182YKV4 A0A182MTA4 A0A182WLM7 A0A182INV4 A0A182US23 A0A182K4J0 A0A182TLD3 A0A182X5E3 Q7Q5W1 A0A182I1A4 A0A182KNV9 A0A023ENX1 A0A182RDU4 A0A182NK13 E1ZZA3 E9ID33 A0A0J7LB80 U5ESN3 F4WVJ1 A0A195BV25 A0A195FL31 A0A158NEG7 A0A151XJC1 A0A195ENC7 A0A1Q3FXR0 A0A1B6CQE3 A0A2M4BVD9 W5JQA3 A0A2M3ZAQ6 A0A0L7QLC8 E0V9Y6 A0A2M4ATR3 A0A195CPJ5 A0A026VTQ5 A0A2A3E1Y7 A0A2R7VPX9 T1JLA9 A0A0N0BK56 A0A2P6KGW6 A0A087SWS9 A0A0C9PVQ9 A0A0P4W0G7 A0A182P7P4 D6X254 A0A2L2XXS7 V5GCA1 A0A232FA97 K7IQ87 A0A1W4XI70 A0A1Y1L2T7 A0A0K8UR19 A0A034VYC4 W8ALV6 A0A1Z5L4W8 A0A293LTF4 A0A131XBK0 L7MES0 A0A224YUQ6 A0A131YWE0 A0A0T6B502 A0A1E1XJ86 A0A3R7PNT1 A0A2R5L7A3 V5HXG5 A0A087ZR51 A0A3S3PJI0 B7PNB0 A0A182FH40 A0A182S6X5 A0A2R7VRX5 B4LPG5 B4J6H1 Q7JZ62 B4KLS8 A0A1A9X4D5 B4GBI0 B4ISY5 B4ILK5 B4QC32 A0A1W4W1M2 A0A1A9VT55
A0A194QMI5 A0A194PE83 S4PFJ4 A0A0L7KWP3 A0A067R7M6 A0A1B6FLY3 A0A1B6J6G6 A0A2J7QTS5 A0A0N7Z9B0 R4G4Q0 A0A224XGY7 A0A023F9T2 E2B9K5 A0A182YKV4 A0A182MTA4 A0A182WLM7 A0A182INV4 A0A182US23 A0A182K4J0 A0A182TLD3 A0A182X5E3 Q7Q5W1 A0A182I1A4 A0A182KNV9 A0A023ENX1 A0A182RDU4 A0A182NK13 E1ZZA3 E9ID33 A0A0J7LB80 U5ESN3 F4WVJ1 A0A195BV25 A0A195FL31 A0A158NEG7 A0A151XJC1 A0A195ENC7 A0A1Q3FXR0 A0A1B6CQE3 A0A2M4BVD9 W5JQA3 A0A2M3ZAQ6 A0A0L7QLC8 E0V9Y6 A0A2M4ATR3 A0A195CPJ5 A0A026VTQ5 A0A2A3E1Y7 A0A2R7VPX9 T1JLA9 A0A0N0BK56 A0A2P6KGW6 A0A087SWS9 A0A0C9PVQ9 A0A0P4W0G7 A0A182P7P4 D6X254 A0A2L2XXS7 V5GCA1 A0A232FA97 K7IQ87 A0A1W4XI70 A0A1Y1L2T7 A0A0K8UR19 A0A034VYC4 W8ALV6 A0A1Z5L4W8 A0A293LTF4 A0A131XBK0 L7MES0 A0A224YUQ6 A0A131YWE0 A0A0T6B502 A0A1E1XJ86 A0A3R7PNT1 A0A2R5L7A3 V5HXG5 A0A087ZR51 A0A3S3PJI0 B7PNB0 A0A182FH40 A0A182S6X5 A0A2R7VRX5 B4LPG5 B4J6H1 Q7JZ62 B4KLS8 A0A1A9X4D5 B4GBI0 B4ISY5 B4ILK5 B4QC32 A0A1W4W1M2 A0A1A9VT55
Pubmed
19121390
22118469
26354079
23622113
26227816
24845553
+ More
27129103 25474469 20798317 25244985 12364791 14747013 17210077 20966253 24945155 26483478 21282665 21719571 21347285 20920257 23761445 20566863 24508170 30249741 18362917 19820115 26561354 28648823 20075255 28004739 25348373 24495485 28528879 28049606 25576852 28797301 26830274 29209593 25765539 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249
27129103 25474469 20798317 25244985 12364791 14747013 17210077 20966253 24945155 26483478 21282665 21719571 21347285 20920257 23761445 20566863 24508170 30249741 18362917 19820115 26561354 28648823 20075255 28004739 25348373 24495485 28528879 28049606 25576852 28797301 26830274 29209593 25765539 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249
EMBL
BABH01008749
NWSH01000455
PCG76298.1
ODYU01009143
SOQ53359.1
GDQN01002478
+ More
JAT88576.1 AGBW02005235 OWR55331.1 RSAL01000073 RVE48996.1 KQ461198 KPJ06165.1 KQ459606 KPI91572.1 GAIX01006560 JAA86000.1 JTDY01004868 KOB67653.1 KK852824 KDR15475.1 GECZ01018563 JAS51206.1 GECU01012943 GECU01004821 JAS94763.1 JAT02886.1 NEVH01011193 PNF31992.1 GDKW01001051 JAI55544.1 ACPB03000375 GAHY01000586 JAA76924.1 GFTR01004669 JAW11757.1 GBBI01000562 JAC18150.1 GL446556 EFN87638.1 AXCM01005227 AAAB01008960 EAA11352.3 APCN01000048 JXUM01052654 GAPW01002947 KQ561744 JAC10651.1 KXJ77654.1 GL435311 EFN73518.1 GL762454 EFZ21472.1 LBMM01000067 KMR05093.1 GANO01003167 JAB56704.1 GL888387 EGI61812.1 KQ976401 KYM92474.1 KQ981490 KYN41123.1 ADTU01013294 KQ982080 KYQ60395.1 KQ978625 KYN29661.1 GFDL01002772 JAV32273.1 GEDC01028358 GEDC01021855 GEDC01012280 JAS08940.1 JAS15443.1 JAS25018.1 GGFJ01007909 MBW57050.1 ADMH02000773 ETN65085.1 GGFM01004852 MBW25603.1 KQ414924 KOC59413.1 DS235000 EEB10192.1 GGFK01010836 MBW44157.1 KQ977444 KYN02658.1 KK107965 QOIP01000001 EZA47065.1 RLU27648.1 KZ288442 PBC25708.1 KK854023 PTY09582.1 JH432107 KQ435709 KOX79998.1 MWRG01012262 PRD25570.1 KK112309 KFM57318.1 GBYB01005553 JAG75320.1 GDRN01097778 JAI59078.1 KQ971371 EFA10215.2 IAAA01008761 IAAA01008762 LAA00804.1 GALX01000691 JAB67775.1 NNAY01000613 OXU27400.1 GEZM01066422 JAV67894.1 GDHF01023170 JAI29144.1 GAKP01010656 GAKP01010653 JAC48296.1 GAMC01016795 GAMC01016792 JAB89760.1 GFJQ02004660 JAW02310.1 GFWV01003062 GFWV01006606 MAA31336.1 GEFH01005043 JAP63538.1 GACK01002707 JAA62327.1 GFPF01006416 MAA17562.1 GEDV01005280 JAP83277.1 LJIG01015985 KRT81959.1 GFAA01004099 JAT99335.1 QCYY01001457 ROT77916.1 GGLE01001181 MBY05307.1 GANP01005240 JAB79228.1 NCKU01000219 RWS16506.1 ABJB010064295 ABJB010103024 ABJB010551634 ABJB010571975 ABJB010598711 DS751891 EEC08082.1 PTY09581.1 CH940648 EDW61224.1 CH916367 CH917341 EDW01972.1 EDW05137.1 AE013599 AY071521 AAF57336.1 AAL49143.1 CH933808 EDW09738.1 CH479181 EDW31275.1 CH891608 EDW99567.1 CH480874 EDW53893.1 CM000362 CM002911 EDX05804.1 KMY91612.1
JAT88576.1 AGBW02005235 OWR55331.1 RSAL01000073 RVE48996.1 KQ461198 KPJ06165.1 KQ459606 KPI91572.1 GAIX01006560 JAA86000.1 JTDY01004868 KOB67653.1 KK852824 KDR15475.1 GECZ01018563 JAS51206.1 GECU01012943 GECU01004821 JAS94763.1 JAT02886.1 NEVH01011193 PNF31992.1 GDKW01001051 JAI55544.1 ACPB03000375 GAHY01000586 JAA76924.1 GFTR01004669 JAW11757.1 GBBI01000562 JAC18150.1 GL446556 EFN87638.1 AXCM01005227 AAAB01008960 EAA11352.3 APCN01000048 JXUM01052654 GAPW01002947 KQ561744 JAC10651.1 KXJ77654.1 GL435311 EFN73518.1 GL762454 EFZ21472.1 LBMM01000067 KMR05093.1 GANO01003167 JAB56704.1 GL888387 EGI61812.1 KQ976401 KYM92474.1 KQ981490 KYN41123.1 ADTU01013294 KQ982080 KYQ60395.1 KQ978625 KYN29661.1 GFDL01002772 JAV32273.1 GEDC01028358 GEDC01021855 GEDC01012280 JAS08940.1 JAS15443.1 JAS25018.1 GGFJ01007909 MBW57050.1 ADMH02000773 ETN65085.1 GGFM01004852 MBW25603.1 KQ414924 KOC59413.1 DS235000 EEB10192.1 GGFK01010836 MBW44157.1 KQ977444 KYN02658.1 KK107965 QOIP01000001 EZA47065.1 RLU27648.1 KZ288442 PBC25708.1 KK854023 PTY09582.1 JH432107 KQ435709 KOX79998.1 MWRG01012262 PRD25570.1 KK112309 KFM57318.1 GBYB01005553 JAG75320.1 GDRN01097778 JAI59078.1 KQ971371 EFA10215.2 IAAA01008761 IAAA01008762 LAA00804.1 GALX01000691 JAB67775.1 NNAY01000613 OXU27400.1 GEZM01066422 JAV67894.1 GDHF01023170 JAI29144.1 GAKP01010656 GAKP01010653 JAC48296.1 GAMC01016795 GAMC01016792 JAB89760.1 GFJQ02004660 JAW02310.1 GFWV01003062 GFWV01006606 MAA31336.1 GEFH01005043 JAP63538.1 GACK01002707 JAA62327.1 GFPF01006416 MAA17562.1 GEDV01005280 JAP83277.1 LJIG01015985 KRT81959.1 GFAA01004099 JAT99335.1 QCYY01001457 ROT77916.1 GGLE01001181 MBY05307.1 GANP01005240 JAB79228.1 NCKU01000219 RWS16506.1 ABJB010064295 ABJB010103024 ABJB010551634 ABJB010571975 ABJB010598711 DS751891 EEC08082.1 PTY09581.1 CH940648 EDW61224.1 CH916367 CH917341 EDW01972.1 EDW05137.1 AE013599 AY071521 AAF57336.1 AAL49143.1 CH933808 EDW09738.1 CH479181 EDW31275.1 CH891608 EDW99567.1 CH480874 EDW53893.1 CM000362 CM002911 EDX05804.1 KMY91612.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000037510 UP000027135 UP000235965 UP000015103 UP000008237 UP000076408 UP000075883 UP000075920 UP000075880 UP000075903 UP000075881 UP000075902 UP000076407 UP000007062 UP000075840 UP000075882 UP000069940 UP000249989 UP000075900 UP000075884 UP000000311 UP000036403 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000078492 UP000000673 UP000053825 UP000009046 UP000078542 UP000053097 UP000279307 UP000242457 UP000053105 UP000054359 UP000075885 UP000007266 UP000215335 UP000002358 UP000192223 UP000283509 UP000005203 UP000285301 UP000001555 UP000069272 UP000075901 UP000008792 UP000001070 UP000000803 UP000009192 UP000091820 UP000008744 UP000002282 UP000001292 UP000000304 UP000192221 UP000078200
UP000037510 UP000027135 UP000235965 UP000015103 UP000008237 UP000076408 UP000075883 UP000075920 UP000075880 UP000075903 UP000075881 UP000075902 UP000076407 UP000007062 UP000075840 UP000075882 UP000069940 UP000249989 UP000075900 UP000075884 UP000000311 UP000036403 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000078492 UP000000673 UP000053825 UP000009046 UP000078542 UP000053097 UP000279307 UP000242457 UP000053105 UP000054359 UP000075885 UP000007266 UP000215335 UP000002358 UP000192223 UP000283509 UP000005203 UP000285301 UP000001555 UP000069272 UP000075901 UP000008792 UP000001070 UP000000803 UP000009192 UP000091820 UP000008744 UP000002282 UP000001292 UP000000304 UP000192221 UP000078200
Interpro
Gene 3D
ProteinModelPortal
H9JAL7
A0A2A4JX72
A0A2H1WJY7
A0A1E1WP15
A0A212FNK9
A0A3S2M1R6
+ More
A0A194QMI5 A0A194PE83 S4PFJ4 A0A0L7KWP3 A0A067R7M6 A0A1B6FLY3 A0A1B6J6G6 A0A2J7QTS5 A0A0N7Z9B0 R4G4Q0 A0A224XGY7 A0A023F9T2 E2B9K5 A0A182YKV4 A0A182MTA4 A0A182WLM7 A0A182INV4 A0A182US23 A0A182K4J0 A0A182TLD3 A0A182X5E3 Q7Q5W1 A0A182I1A4 A0A182KNV9 A0A023ENX1 A0A182RDU4 A0A182NK13 E1ZZA3 E9ID33 A0A0J7LB80 U5ESN3 F4WVJ1 A0A195BV25 A0A195FL31 A0A158NEG7 A0A151XJC1 A0A195ENC7 A0A1Q3FXR0 A0A1B6CQE3 A0A2M4BVD9 W5JQA3 A0A2M3ZAQ6 A0A0L7QLC8 E0V9Y6 A0A2M4ATR3 A0A195CPJ5 A0A026VTQ5 A0A2A3E1Y7 A0A2R7VPX9 T1JLA9 A0A0N0BK56 A0A2P6KGW6 A0A087SWS9 A0A0C9PVQ9 A0A0P4W0G7 A0A182P7P4 D6X254 A0A2L2XXS7 V5GCA1 A0A232FA97 K7IQ87 A0A1W4XI70 A0A1Y1L2T7 A0A0K8UR19 A0A034VYC4 W8ALV6 A0A1Z5L4W8 A0A293LTF4 A0A131XBK0 L7MES0 A0A224YUQ6 A0A131YWE0 A0A0T6B502 A0A1E1XJ86 A0A3R7PNT1 A0A2R5L7A3 V5HXG5 A0A087ZR51 A0A3S3PJI0 B7PNB0 A0A182FH40 A0A182S6X5 A0A2R7VRX5 B4LPG5 B4J6H1 Q7JZ62 B4KLS8 A0A1A9X4D5 B4GBI0 B4ISY5 B4ILK5 B4QC32 A0A1W4W1M2 A0A1A9VT55
A0A194QMI5 A0A194PE83 S4PFJ4 A0A0L7KWP3 A0A067R7M6 A0A1B6FLY3 A0A1B6J6G6 A0A2J7QTS5 A0A0N7Z9B0 R4G4Q0 A0A224XGY7 A0A023F9T2 E2B9K5 A0A182YKV4 A0A182MTA4 A0A182WLM7 A0A182INV4 A0A182US23 A0A182K4J0 A0A182TLD3 A0A182X5E3 Q7Q5W1 A0A182I1A4 A0A182KNV9 A0A023ENX1 A0A182RDU4 A0A182NK13 E1ZZA3 E9ID33 A0A0J7LB80 U5ESN3 F4WVJ1 A0A195BV25 A0A195FL31 A0A158NEG7 A0A151XJC1 A0A195ENC7 A0A1Q3FXR0 A0A1B6CQE3 A0A2M4BVD9 W5JQA3 A0A2M3ZAQ6 A0A0L7QLC8 E0V9Y6 A0A2M4ATR3 A0A195CPJ5 A0A026VTQ5 A0A2A3E1Y7 A0A2R7VPX9 T1JLA9 A0A0N0BK56 A0A2P6KGW6 A0A087SWS9 A0A0C9PVQ9 A0A0P4W0G7 A0A182P7P4 D6X254 A0A2L2XXS7 V5GCA1 A0A232FA97 K7IQ87 A0A1W4XI70 A0A1Y1L2T7 A0A0K8UR19 A0A034VYC4 W8ALV6 A0A1Z5L4W8 A0A293LTF4 A0A131XBK0 L7MES0 A0A224YUQ6 A0A131YWE0 A0A0T6B502 A0A1E1XJ86 A0A3R7PNT1 A0A2R5L7A3 V5HXG5 A0A087ZR51 A0A3S3PJI0 B7PNB0 A0A182FH40 A0A182S6X5 A0A2R7VRX5 B4LPG5 B4J6H1 Q7JZ62 B4KLS8 A0A1A9X4D5 B4GBI0 B4ISY5 B4ILK5 B4QC32 A0A1W4W1M2 A0A1A9VT55
PDB
5FTA
E-value=2.79623e-38,
Score=397
Ontologies
GO
Topology
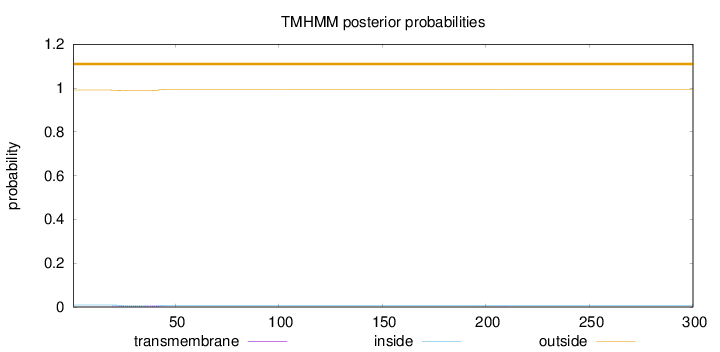
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1302
Exp number, first 60 AAs:
0.1276
Total prob of N-in:
0.00932
outside
1 - 300
Population Genetic Test Statistics
Pi
203.326068
Theta
178.618625
Tajima's D
0.459406
CLR
0.023624
CSRT
0.503474826258687
Interpretation
Uncertain
