Gene
KWMTBOMO05617
Pre Gene Modal
BGIBMGA006776
Annotation
PREDICTED:_heparan_sulfate_glucosamine_3-O-sulfotransferase_5_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.386 Nuclear Reliability : 2.045
Sequence
CDS
ATGAGTCAGTTGCCGAATAAGCACCATCGCCCTGAGGCGACTTACAGAAGTTGTTCTCAGTCCTTGCCTCTTGATATCCTATGGTTGCCGCGGTCGTCGTTCCGGCCGGCGGAGGGCGAGCTGGCGGACTGCGTTCTGGTGGTGGGCGTGTCGCGGCCCAAGCTAGCCGCGGCGCTGCTCTCCGTCACGCTACTGTCGCTCTTTCTCACCTTCCATGTTCTCTATGACAGCGCACTCTATAGCATACAGGCGGCCAGCATGGCGTCGGCTGCTAGCGAGAATCGCAAGATGTTGCTTAGCCAAGAGAGTGACACGCGTACCATCAGTCACCCCATGGCGTTACGGAAGAGACTTAGGCCTCCTAAGCAGCCTAGACCTGCTCGAAGACTTCCACAGGCTTTAATCATCGGCGTCCGGAAATGTGGCACTCGAGCTCTCCTGGAAATGCTGTACCTACATCCGATGGTCCAAAAGGCTTCCGGAGAAGTTCATTTCTTCGACAGAGATGAGAACTACGCTTTGGGCCTGGAATGGTACAGAAGTAAAATGCCGTTGTCTTTCAAAGGACAAATCACCATTGAGAAGAGTCCTAGCTATTTTGTTACGCCAGAGGTAAGGGCAACCGAAAGGATACGTTAA
Protein
MSQLPNKHHRPEATYRSCSQSLPLDILWLPRSSFRPAEGELADCVLVVGVSRPKLAAALLSVTLLSLFLTFHVLYDSALYSIQAASMASAASENRKMLLSQESDTRTISHPMALRKRLRPPKQPRPARRLPQALIIGVRKCGTRALLEMLYLHPMVQKASGEVHFFDRDENYALGLEWYRSKMPLSFKGQITIEKSPSYFVTPEVRATERIR
Summary
Uniprot
A0A2A4J1S8
A0A194QKV9
A0A212ESY3
A0A2H1WJ11
A0A194PFU8
A0A0L7KUX9
+ More
W5JLS3 A0A182Q703 A0A1Y1LXL8 A0A3L8E313 A0A2M4CWJ5 A0A087ZR12 A0A2M4CXP6 A0A2M4CWD8 A0A158NEG2 B4KPK5 A0A0T6BDG8 B4MK07 D6WZI2 B4LJ39 B3MI27 A0A195EN96 A0A0M4EHK3 Q8MRE7 W8B7Y9 A0A195CPV1 A0A026VTJ2 A0A1A9XHB1 A0A182X5D9 A0A182RDU0 A0A0N0BK33 A0A182TSM6 A0A182M0T7 A0A0L7QLD5 A0A182V7U8 A0A182KNW1 A0A154P5P7 A0A182P7P8 A0A182IYR1 A0A2M4AXB5 A0A195FL99 E9ID31 A0A182GA57 A0A182YKV0 A0A310SNV9 E2B9K3 A0A182NK17 F4WVJ3 U4U7K9 E1ZZA7 A0A067QYP7 K7IRQ7 A0A084WL20 A0A182FH45 A0A2J7QTU5 A0A1J1JA79 A0A1J1J697 A0A2P8ZAI9 A0A1B0D339
W5JLS3 A0A182Q703 A0A1Y1LXL8 A0A3L8E313 A0A2M4CWJ5 A0A087ZR12 A0A2M4CXP6 A0A2M4CWD8 A0A158NEG2 B4KPK5 A0A0T6BDG8 B4MK07 D6WZI2 B4LJ39 B3MI27 A0A195EN96 A0A0M4EHK3 Q8MRE7 W8B7Y9 A0A195CPV1 A0A026VTJ2 A0A1A9XHB1 A0A182X5D9 A0A182RDU0 A0A0N0BK33 A0A182TSM6 A0A182M0T7 A0A0L7QLD5 A0A182V7U8 A0A182KNW1 A0A154P5P7 A0A182P7P8 A0A182IYR1 A0A2M4AXB5 A0A195FL99 E9ID31 A0A182GA57 A0A182YKV0 A0A310SNV9 E2B9K3 A0A182NK17 F4WVJ3 U4U7K9 E1ZZA7 A0A067QYP7 K7IRQ7 A0A084WL20 A0A182FH45 A0A2J7QTU5 A0A1J1JA79 A0A1J1J697 A0A2P8ZAI9 A0A1B0D339
Pubmed
26354079
22118469
26227816
20920257
23761445
28004739
+ More
30249741 21347285 17994087 18362917 19820115 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 24508170 20966253 21282665 26483478 25244985 20798317 21719571 23537049 24845553 20075255 24438588 29403074
30249741 21347285 17994087 18362917 19820115 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 24508170 20966253 21282665 26483478 25244985 20798317 21719571 23537049 24845553 20075255 24438588 29403074
EMBL
NWSH01003555
PCG66117.1
KQ461198
KPJ06167.1
AGBW02012691
OWR44605.1
+ More
ODYU01008972 SOQ53017.1 KQ459606 KPI91574.1 JTDY01005378 KOB67062.1 ADMH02000773 ETN65081.1 AXCN02000496 GEZM01045939 JAV77608.1 QOIP01000001 RLU27036.1 GGFL01005461 MBW69639.1 GGFL01005460 MBW69638.1 GGFL01005462 MBW69640.1 ADTU01013289 CH933808 EDW09115.2 LJIG01001554 KRT85386.1 CH963846 EDW72446.1 KQ971372 EFA09701.2 CH940648 EDW61475.2 CH902619 EDV38037.1 KPU77227.1 KPU77228.1 KPU77229.1 KPU77230.1 KQ978625 KYN29663.1 CP012524 ALC42072.1 AE013599 AY121626 AAF57644.2 AAM51953.1 AHN56384.1 GAMC01011828 JAB94727.1 KQ977444 KYN02660.1 KK107965 EZA47062.1 KQ435709 KOX79992.1 AXCM01003374 KQ414924 KOC59420.1 KQ434822 KZC07197.1 GGFK01012099 MBW45420.1 KQ981490 KYN41121.1 GL762454 EFZ21402.1 JXUM01008586 JXUM01008587 JXUM01008588 JXUM01008589 JXUM01008590 JXUM01008591 JXUM01008592 JXUM01008593 JXUM01008594 KQ560264 KXJ83477.1 KQ759804 OAD62792.1 GL446556 EFN87636.1 GL888387 EGI61814.1 KB632191 ERL89869.1 GL435311 EFN73522.1 KK852824 KDR15472.1 ATLV01024170 KE525350 KFB50914.1 NEVH01011193 PNF31995.1 CVRI01000074 CRL07929.1 CRL07931.1 PYGN01000124 PSN53510.1 AJVK01023234 AJVK01023235
ODYU01008972 SOQ53017.1 KQ459606 KPI91574.1 JTDY01005378 KOB67062.1 ADMH02000773 ETN65081.1 AXCN02000496 GEZM01045939 JAV77608.1 QOIP01000001 RLU27036.1 GGFL01005461 MBW69639.1 GGFL01005460 MBW69638.1 GGFL01005462 MBW69640.1 ADTU01013289 CH933808 EDW09115.2 LJIG01001554 KRT85386.1 CH963846 EDW72446.1 KQ971372 EFA09701.2 CH940648 EDW61475.2 CH902619 EDV38037.1 KPU77227.1 KPU77228.1 KPU77229.1 KPU77230.1 KQ978625 KYN29663.1 CP012524 ALC42072.1 AE013599 AY121626 AAF57644.2 AAM51953.1 AHN56384.1 GAMC01011828 JAB94727.1 KQ977444 KYN02660.1 KK107965 EZA47062.1 KQ435709 KOX79992.1 AXCM01003374 KQ414924 KOC59420.1 KQ434822 KZC07197.1 GGFK01012099 MBW45420.1 KQ981490 KYN41121.1 GL762454 EFZ21402.1 JXUM01008586 JXUM01008587 JXUM01008588 JXUM01008589 JXUM01008590 JXUM01008591 JXUM01008592 JXUM01008593 JXUM01008594 KQ560264 KXJ83477.1 KQ759804 OAD62792.1 GL446556 EFN87636.1 GL888387 EGI61814.1 KB632191 ERL89869.1 GL435311 EFN73522.1 KK852824 KDR15472.1 ATLV01024170 KE525350 KFB50914.1 NEVH01011193 PNF31995.1 CVRI01000074 CRL07929.1 CRL07931.1 PYGN01000124 PSN53510.1 AJVK01023234 AJVK01023235
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
UP000000673
+ More
UP000075886 UP000279307 UP000005203 UP000005205 UP000009192 UP000007798 UP000007266 UP000008792 UP000007801 UP000078492 UP000092553 UP000000803 UP000078542 UP000053097 UP000092443 UP000076407 UP000075900 UP000053105 UP000075902 UP000075883 UP000053825 UP000075903 UP000075882 UP000076502 UP000075885 UP000075880 UP000078541 UP000069940 UP000249989 UP000076408 UP000008237 UP000075884 UP000007755 UP000030742 UP000000311 UP000027135 UP000002358 UP000030765 UP000069272 UP000235965 UP000183832 UP000245037 UP000092462
UP000075886 UP000279307 UP000005203 UP000005205 UP000009192 UP000007798 UP000007266 UP000008792 UP000007801 UP000078492 UP000092553 UP000000803 UP000078542 UP000053097 UP000092443 UP000076407 UP000075900 UP000053105 UP000075902 UP000075883 UP000053825 UP000075903 UP000075882 UP000076502 UP000075885 UP000075880 UP000078541 UP000069940 UP000249989 UP000076408 UP000008237 UP000075884 UP000007755 UP000030742 UP000000311 UP000027135 UP000002358 UP000030765 UP000069272 UP000235965 UP000183832 UP000245037 UP000092462
PRIDE
Pfam
PF00685 Sulfotransfer_1
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4J1S8
A0A194QKV9
A0A212ESY3
A0A2H1WJ11
A0A194PFU8
A0A0L7KUX9
+ More
W5JLS3 A0A182Q703 A0A1Y1LXL8 A0A3L8E313 A0A2M4CWJ5 A0A087ZR12 A0A2M4CXP6 A0A2M4CWD8 A0A158NEG2 B4KPK5 A0A0T6BDG8 B4MK07 D6WZI2 B4LJ39 B3MI27 A0A195EN96 A0A0M4EHK3 Q8MRE7 W8B7Y9 A0A195CPV1 A0A026VTJ2 A0A1A9XHB1 A0A182X5D9 A0A182RDU0 A0A0N0BK33 A0A182TSM6 A0A182M0T7 A0A0L7QLD5 A0A182V7U8 A0A182KNW1 A0A154P5P7 A0A182P7P8 A0A182IYR1 A0A2M4AXB5 A0A195FL99 E9ID31 A0A182GA57 A0A182YKV0 A0A310SNV9 E2B9K3 A0A182NK17 F4WVJ3 U4U7K9 E1ZZA7 A0A067QYP7 K7IRQ7 A0A084WL20 A0A182FH45 A0A2J7QTU5 A0A1J1JA79 A0A1J1J697 A0A2P8ZAI9 A0A1B0D339
W5JLS3 A0A182Q703 A0A1Y1LXL8 A0A3L8E313 A0A2M4CWJ5 A0A087ZR12 A0A2M4CXP6 A0A2M4CWD8 A0A158NEG2 B4KPK5 A0A0T6BDG8 B4MK07 D6WZI2 B4LJ39 B3MI27 A0A195EN96 A0A0M4EHK3 Q8MRE7 W8B7Y9 A0A195CPV1 A0A026VTJ2 A0A1A9XHB1 A0A182X5D9 A0A182RDU0 A0A0N0BK33 A0A182TSM6 A0A182M0T7 A0A0L7QLD5 A0A182V7U8 A0A182KNW1 A0A154P5P7 A0A182P7P8 A0A182IYR1 A0A2M4AXB5 A0A195FL99 E9ID31 A0A182GA57 A0A182YKV0 A0A310SNV9 E2B9K3 A0A182NK17 F4WVJ3 U4U7K9 E1ZZA7 A0A067QYP7 K7IRQ7 A0A084WL20 A0A182FH45 A0A2J7QTU5 A0A1J1JA79 A0A1J1J697 A0A2P8ZAI9 A0A1B0D339
PDB
3BD9
E-value=3.89744e-25,
Score=281
Ontologies
GO
PANTHER
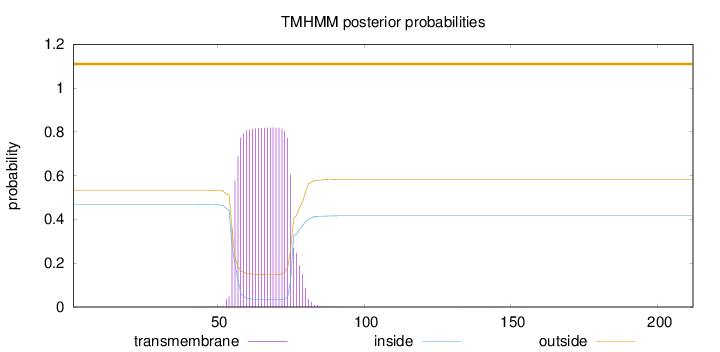
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.1299
Exp number, first 60 AAs:
4.10384
Total prob of N-in:
0.46810
outside
1 - 212
Population Genetic Test Statistics
Pi
274.101455
Theta
166.932633
Tajima's D
2.362059
CLR
0.370951
CSRT
0.931903404829759
Interpretation
Uncertain
