Pre Gene Modal
BGIBMGA006774
Annotation
PREDICTED:_cytochrome_b_reductase_1-like_isoform_X2_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.989
Sequence
CDS
ATGGGTGGACGAGCTCACAGCGCAAATGCTGTTAAGAGGTTACCGGAGCACATAGACATCTACATAGTAAATGCCGCCACCCACCTTGAGATAAAAGTTCTAAGCCAACGCAGTCGGTCCTATGACTCGGACGACCACGAGCGAGGTGCCTGCGAGTGGTGCGAGTATGCCCTCGTGATCACTTTGGCGACGGTGCTGCTCATTGGCGTCTCCGCTCTGACCATCTTCTGGGTGTTCTATTACCGCGGGGGCTATGCCTGGGCCGACGACACCCCGGACAAGCAGTTCAACTTGCACCCGACTCTTATGGTGGCCGGTTTCGTTACATTCTGCGGTTTCTCCGTGCTCTTGTACCGCATCTGTCGATGCTTCCGGCGCATCTATGTCAAACTGCTGCACGCCATCTTCCACGCGTTAGCGTTCCCGTGCATCGTTATCGGCTTCCTGGCTGTCCTCGACTACCACAACAAGAAGGGCATCAACAACTTCTATTCGCTCCACAGCTGGATTGGCTTCGTCGCCATGGGACTGTTTGGATTACAGTACGCAGTCGGCTTCTTCAGCTTCCTCCTACTGTTGATTTGCAATAAAGGCACCGCTGGATTCCGCGCTTCCTTGGTACCGATCCACGCCGCCTTTGGAATCCTGACCTTCGTACTCGGTGTCGCTGCTTGTCTCACCGGACTCACTGAAAAGGCAATCTTTGCCCTCGGGAGCGAGCAGTACCCGCTTTTGGAACCTGAAGCGATCGTTATCAACGCTATCGGAATGACTGTGGTTGCTGTTGCCATCGTTGTGCTTTACACTGTACGCGGCACTGGCGGCTACAGGCTCCGCAAGGTCGCCCAGCACCCGAGTTAA
Protein
MGGRAHSANAVKRLPEHIDIYIVNAATHLEIKVLSQRSRSYDSDDHERGACEWCEYALVITLATVLLIGVSALTIFWVFYYRGGYAWADDTPDKQFNLHPTLMVAGFVTFCGFSVLLYRICRCFRRIYVKLLHAIFHALAFPCIVIGFLAVLDYHNKKGINNFYSLHSWIGFVAMGLFGLQYAVGFFSFLLLLICNKGTAGFRASLVPIHAAFGILTFVLGVAACLTGLTEKAIFALGSEQYPLLEPEAIVINAIGMTVVAVAIVVLYTVRGTGGYRLRKVAQHPS
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RsmB/NOP family.
Uniprot
H9JB79
A0A2A4JSG5
A0A194QKW4
A0A2H1WJH2
A0A212EPY8
I4DML1
+ More
A0A2A4JQS4 A0A2A4JSM8 A0A2A4JS07 I4DL20 S4PVU3 A0A194PFV3 A0A1Y1MNC5 A0A146KMU1 A0A1Y1MND0 A0A069DXX6 A0A023F983 R4G5W5 A0A1W4XT88 A0A1W4XSB1 A0A139WBJ9 D6WZ69 A0A1W4XTD0 A0A1B6DSN9 E0VVT2 A0A1B0C871 A0A1B6CL74 A0A1L8E0H6 A0A1L8DZV3 N6UP03 U4UPS6 A0A182J656 A0A182P8T7 A0A2M4CS21 A0A2M4BX83 A0A2M4BXI1 B4J8F5 A0A0Q9VYR3 B4MFP3 A0A1B6F544 A0A182MFN3 A0A0Q9W5V8 A0A1B6FLN8 A0A0Q9X6Z3 A0A182SQE4 A0A182YC30 A7UTA7 A0A182HNX1 A0A182RKW5 A0A182WK16 A0A1Y9GI20 A0A182XEU4 Q7Q9I3 A0A1B0GBY5 Q16QH9 A0A1S4FSU8 A0A2M3Z8X0 A0A182FCX1 A0A023ELD3 A0A034VHS0 W5JU50 A0A0A1XR97 B4KRS6 A0A0K8W9T2 A0A034VGS4 A0A0Q9XIG3 A0A0A1WK18 A0A0Q9X7T9 A0A0K8U9R1 A0A034VID8 A0A182Q9T3 A0A1S3DCB8 A0A1D6ZM26 A0A0M4EEC4 A0A182U4V9 T1PHK3 A0A1I8PNT6 A0A1I8PNP7 A0A1I8M9F2 A0A1A9ZYR6 A0A1B0BY62 A0A1I8M9F1 A0A1I8PNT9 A0A1W4VQZ1 T1PKG5 A0A0N8P0C5 A0A3B0JM45 A0A0P8XQ64 N6W627 A0A1I8M9E9 A0A3B0JMS7 A0A1W4VD81 C0PTW4 A0A0R3NV20 B3MCU8
A0A2A4JQS4 A0A2A4JSM8 A0A2A4JS07 I4DL20 S4PVU3 A0A194PFV3 A0A1Y1MNC5 A0A146KMU1 A0A1Y1MND0 A0A069DXX6 A0A023F983 R4G5W5 A0A1W4XT88 A0A1W4XSB1 A0A139WBJ9 D6WZ69 A0A1W4XTD0 A0A1B6DSN9 E0VVT2 A0A1B0C871 A0A1B6CL74 A0A1L8E0H6 A0A1L8DZV3 N6UP03 U4UPS6 A0A182J656 A0A182P8T7 A0A2M4CS21 A0A2M4BX83 A0A2M4BXI1 B4J8F5 A0A0Q9VYR3 B4MFP3 A0A1B6F544 A0A182MFN3 A0A0Q9W5V8 A0A1B6FLN8 A0A0Q9X6Z3 A0A182SQE4 A0A182YC30 A7UTA7 A0A182HNX1 A0A182RKW5 A0A182WK16 A0A1Y9GI20 A0A182XEU4 Q7Q9I3 A0A1B0GBY5 Q16QH9 A0A1S4FSU8 A0A2M3Z8X0 A0A182FCX1 A0A023ELD3 A0A034VHS0 W5JU50 A0A0A1XR97 B4KRS6 A0A0K8W9T2 A0A034VGS4 A0A0Q9XIG3 A0A0A1WK18 A0A0Q9X7T9 A0A0K8U9R1 A0A034VID8 A0A182Q9T3 A0A1S3DCB8 A0A1D6ZM26 A0A0M4EEC4 A0A182U4V9 T1PHK3 A0A1I8PNT6 A0A1I8PNP7 A0A1I8M9F2 A0A1A9ZYR6 A0A1B0BY62 A0A1I8M9F1 A0A1I8PNT9 A0A1W4VQZ1 T1PKG5 A0A0N8P0C5 A0A3B0JM45 A0A0P8XQ64 N6W627 A0A1I8M9E9 A0A3B0JMS7 A0A1W4VD81 C0PTW4 A0A0R3NV20 B3MCU8
Pubmed
EMBL
BABH01008727
NWSH01000751
PCG74403.1
KQ461198
KPJ06172.1
ODYU01009072
+ More
SOQ53225.1 AGBW02013342 OWR43562.1 AK402529 BAM19151.1 PCG74405.1 PCG74402.1 PCG74404.1 AK401988 BAM18610.1 GAIX01007938 JAA84622.1 KQ459606 KPI91579.1 GEZM01026197 JAV87161.1 GDHC01020871 JAP97757.1 GEZM01026196 JAV87162.1 GBGD01002670 JAC86219.1 GBBI01000650 JAC18062.1 GAHY01000165 JAA77345.1 KQ971372 KYB25307.1 EFA10387.2 GEDC01025328 GEDC01008601 JAS11970.1 JAS28697.1 DS235813 EEB17488.1 AJWK01000228 AJWK01000229 GEDC01023183 GEDC01013974 JAS14115.1 JAS23324.1 GFDF01002089 JAV11995.1 GFDF01002061 JAV12023.1 APGK01002698 APGK01002699 KB734918 ENN83480.1 KB632312 ERL92151.1 GGFL01003783 MBW67961.1 GGFJ01008400 MBW57541.1 GGFJ01008635 MBW57776.1 CH916367 EDW02314.1 CH940667 KRF77670.1 EDW57214.1 GECZ01024474 GECZ01015662 GECZ01009957 JAS45295.1 JAS54107.1 JAS59812.1 AXCM01011146 KRF77671.1 GECZ01018667 GECZ01014382 JAS51102.1 JAS55387.1 CH933808 KRG04070.1 AAAB01008900 EDO64114.1 APCN01002800 APCN01002801 APCN01002802 EAA09405.2 CCAG010005902 CH477747 EAT36661.1 GGFM01004194 MBW24945.1 GAPW01003572 GEHC01000229 JAC10026.1 JAV47416.1 GAKP01017642 JAC41310.1 ADMH02000362 ETN66823.1 GBXI01001189 JAD13103.1 EDW08346.1 GDHF01004482 JAI47832.1 GAKP01017645 JAC41307.1 KRG04068.1 KRG04071.1 GBXI01015554 JAC98737.1 KRG04069.1 GDHF01028996 GDHF01003706 JAI23318.1 JAI48608.1 GAKP01017647 GAKP01017644 JAC41308.1 AXCN02001019 KT722698 ANS71633.1 CP012524 ALC41501.1 KA648302 AFP62931.1 JXJN01022490 KA647945 KA649194 AFP63823.1 CH902619 KPU76740.1 OUUW01000001 SPP74619.1 KPU76739.1 CM000071 ENO01708.2 SPP74616.1 BT071809 ACN58572.1 KRT02627.1 EDV37350.1
SOQ53225.1 AGBW02013342 OWR43562.1 AK402529 BAM19151.1 PCG74405.1 PCG74402.1 PCG74404.1 AK401988 BAM18610.1 GAIX01007938 JAA84622.1 KQ459606 KPI91579.1 GEZM01026197 JAV87161.1 GDHC01020871 JAP97757.1 GEZM01026196 JAV87162.1 GBGD01002670 JAC86219.1 GBBI01000650 JAC18062.1 GAHY01000165 JAA77345.1 KQ971372 KYB25307.1 EFA10387.2 GEDC01025328 GEDC01008601 JAS11970.1 JAS28697.1 DS235813 EEB17488.1 AJWK01000228 AJWK01000229 GEDC01023183 GEDC01013974 JAS14115.1 JAS23324.1 GFDF01002089 JAV11995.1 GFDF01002061 JAV12023.1 APGK01002698 APGK01002699 KB734918 ENN83480.1 KB632312 ERL92151.1 GGFL01003783 MBW67961.1 GGFJ01008400 MBW57541.1 GGFJ01008635 MBW57776.1 CH916367 EDW02314.1 CH940667 KRF77670.1 EDW57214.1 GECZ01024474 GECZ01015662 GECZ01009957 JAS45295.1 JAS54107.1 JAS59812.1 AXCM01011146 KRF77671.1 GECZ01018667 GECZ01014382 JAS51102.1 JAS55387.1 CH933808 KRG04070.1 AAAB01008900 EDO64114.1 APCN01002800 APCN01002801 APCN01002802 EAA09405.2 CCAG010005902 CH477747 EAT36661.1 GGFM01004194 MBW24945.1 GAPW01003572 GEHC01000229 JAC10026.1 JAV47416.1 GAKP01017642 JAC41310.1 ADMH02000362 ETN66823.1 GBXI01001189 JAD13103.1 EDW08346.1 GDHF01004482 JAI47832.1 GAKP01017645 JAC41307.1 KRG04068.1 KRG04071.1 GBXI01015554 JAC98737.1 KRG04069.1 GDHF01028996 GDHF01003706 JAI23318.1 JAI48608.1 GAKP01017647 GAKP01017644 JAC41308.1 AXCN02001019 KT722698 ANS71633.1 CP012524 ALC41501.1 KA648302 AFP62931.1 JXJN01022490 KA647945 KA649194 AFP63823.1 CH902619 KPU76740.1 OUUW01000001 SPP74619.1 KPU76739.1 CM000071 ENO01708.2 SPP74616.1 BT071809 ACN58572.1 KRT02627.1 EDV37350.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000192223
+ More
UP000007266 UP000009046 UP000092461 UP000019118 UP000030742 UP000075880 UP000075885 UP000001070 UP000008792 UP000075883 UP000009192 UP000075901 UP000076408 UP000007062 UP000075840 UP000075900 UP000075920 UP000076407 UP000092444 UP000008820 UP000069272 UP000000673 UP000075886 UP000079169 UP000092553 UP000075902 UP000095300 UP000095301 UP000092445 UP000092460 UP000192221 UP000007801 UP000268350 UP000001819
UP000007266 UP000009046 UP000092461 UP000019118 UP000030742 UP000075880 UP000075885 UP000001070 UP000008792 UP000075883 UP000009192 UP000075901 UP000076408 UP000007062 UP000075840 UP000075900 UP000075920 UP000076407 UP000092444 UP000008820 UP000069272 UP000000673 UP000075886 UP000079169 UP000092553 UP000075902 UP000095300 UP000095301 UP000092445 UP000092460 UP000192221 UP000007801 UP000268350 UP000001819
Interpro
ProteinModelPortal
H9JB79
A0A2A4JSG5
A0A194QKW4
A0A2H1WJH2
A0A212EPY8
I4DML1
+ More
A0A2A4JQS4 A0A2A4JSM8 A0A2A4JS07 I4DL20 S4PVU3 A0A194PFV3 A0A1Y1MNC5 A0A146KMU1 A0A1Y1MND0 A0A069DXX6 A0A023F983 R4G5W5 A0A1W4XT88 A0A1W4XSB1 A0A139WBJ9 D6WZ69 A0A1W4XTD0 A0A1B6DSN9 E0VVT2 A0A1B0C871 A0A1B6CL74 A0A1L8E0H6 A0A1L8DZV3 N6UP03 U4UPS6 A0A182J656 A0A182P8T7 A0A2M4CS21 A0A2M4BX83 A0A2M4BXI1 B4J8F5 A0A0Q9VYR3 B4MFP3 A0A1B6F544 A0A182MFN3 A0A0Q9W5V8 A0A1B6FLN8 A0A0Q9X6Z3 A0A182SQE4 A0A182YC30 A7UTA7 A0A182HNX1 A0A182RKW5 A0A182WK16 A0A1Y9GI20 A0A182XEU4 Q7Q9I3 A0A1B0GBY5 Q16QH9 A0A1S4FSU8 A0A2M3Z8X0 A0A182FCX1 A0A023ELD3 A0A034VHS0 W5JU50 A0A0A1XR97 B4KRS6 A0A0K8W9T2 A0A034VGS4 A0A0Q9XIG3 A0A0A1WK18 A0A0Q9X7T9 A0A0K8U9R1 A0A034VID8 A0A182Q9T3 A0A1S3DCB8 A0A1D6ZM26 A0A0M4EEC4 A0A182U4V9 T1PHK3 A0A1I8PNT6 A0A1I8PNP7 A0A1I8M9F2 A0A1A9ZYR6 A0A1B0BY62 A0A1I8M9F1 A0A1I8PNT9 A0A1W4VQZ1 T1PKG5 A0A0N8P0C5 A0A3B0JM45 A0A0P8XQ64 N6W627 A0A1I8M9E9 A0A3B0JMS7 A0A1W4VD81 C0PTW4 A0A0R3NV20 B3MCU8
A0A2A4JQS4 A0A2A4JSM8 A0A2A4JS07 I4DL20 S4PVU3 A0A194PFV3 A0A1Y1MNC5 A0A146KMU1 A0A1Y1MND0 A0A069DXX6 A0A023F983 R4G5W5 A0A1W4XT88 A0A1W4XSB1 A0A139WBJ9 D6WZ69 A0A1W4XTD0 A0A1B6DSN9 E0VVT2 A0A1B0C871 A0A1B6CL74 A0A1L8E0H6 A0A1L8DZV3 N6UP03 U4UPS6 A0A182J656 A0A182P8T7 A0A2M4CS21 A0A2M4BX83 A0A2M4BXI1 B4J8F5 A0A0Q9VYR3 B4MFP3 A0A1B6F544 A0A182MFN3 A0A0Q9W5V8 A0A1B6FLN8 A0A0Q9X6Z3 A0A182SQE4 A0A182YC30 A7UTA7 A0A182HNX1 A0A182RKW5 A0A182WK16 A0A1Y9GI20 A0A182XEU4 Q7Q9I3 A0A1B0GBY5 Q16QH9 A0A1S4FSU8 A0A2M3Z8X0 A0A182FCX1 A0A023ELD3 A0A034VHS0 W5JU50 A0A0A1XR97 B4KRS6 A0A0K8W9T2 A0A034VGS4 A0A0Q9XIG3 A0A0A1WK18 A0A0Q9X7T9 A0A0K8U9R1 A0A034VID8 A0A182Q9T3 A0A1S3DCB8 A0A1D6ZM26 A0A0M4EEC4 A0A182U4V9 T1PHK3 A0A1I8PNT6 A0A1I8PNP7 A0A1I8M9F2 A0A1A9ZYR6 A0A1B0BY62 A0A1I8M9F1 A0A1I8PNT9 A0A1W4VQZ1 T1PKG5 A0A0N8P0C5 A0A3B0JM45 A0A0P8XQ64 N6W627 A0A1I8M9E9 A0A3B0JMS7 A0A1W4VD81 C0PTW4 A0A0R3NV20 B3MCU8
PDB
5ZLG
E-value=5.13065e-29,
Score=316
Ontologies
GO
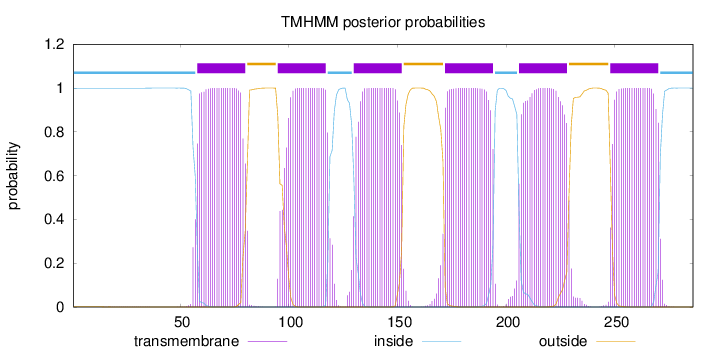
Topology
Length:
286
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
135.47367
Exp number, first 60 AAs:
3.41267
Total prob of N-in:
0.99925
inside
1 - 57
TMhelix
58 - 80
outside
81 - 94
TMhelix
95 - 117
inside
118 - 129
TMhelix
130 - 152
outside
153 - 171
TMhelix
172 - 194
inside
195 - 205
TMhelix
206 - 228
outside
229 - 247
TMhelix
248 - 270
inside
271 - 286
Population Genetic Test Statistics
Pi
237.899943
Theta
200.985673
Tajima's D
1.096568
CLR
0.333756
CSRT
0.68301584920754
Interpretation
Uncertain
