Gene
KWMTBOMO05611
Pre Gene Modal
BGIBMGA006568
Annotation
Uncharacterized_protein_OBRU01_09759_[Operophtera_brumata]
Full name
Coenzyme Q-binding protein COQ10 homolog A, mitochondrial
Location in the cell
Extracellular Reliability : 1.042 Mitochondrial Reliability : 1.46
Sequence
CDS
ATGATTATCTCTCAAAGTTGTGCCGTAGGGCGATACATTTTTCGCAAGAAAAATGTAGTTTTCAGTTTTCAGGGTCATTACGTAAATAGACAAGGTAGTGCACTATGTCATTGCCAAGGCATTCAACAACAAAATCGTAGTTTTATAAACCTGCCAATAACAAACAAAACAAGAGTTTATACTGGAAGGCAACTTGTTGGCTACACAATGGAGCAAATGTTTGAAGTTGTATCAGATGTAGGAAGCTACAACAAATTCCTACCGTGGTGCAAAAAGTCTATAGTTTTGAAAGAAACACCAGGAAACCTGAAGGCGGACTTGATCATTGGCTTTCCTCCTATCAATGAAAGCTACACTTCAAATGTTACGCTAGTCAAACCACATTTGGTAAAAGCAGAGTGTTCTGATGGAAGACTCTTCCACCATATGTTGACCCTTTGGAGATTCAGTCCTGGTCTAAAAAGAGAACAGCAGTCGTGTGTTGTGGACTTCCAGATAACATTCGAATTTAGATCAGCTATACATTCACATTTATCTAATTTGTTTTTTGACCAAGTTGCTCGACAAATGGAAGGTGCTTTCATTAAAGAAGTCGGAAGGCGCAATGGGCCGGCAACAATGCAGCCTAGAAAGTTACTAATAAACAATCACACTTTGAAAACTTGA
Protein
MIISQSCAVGRYIFRKKNVVFSFQGHYVNRQGSALCHCQGIQQQNRSFINLPITNKTRVYTGRQLVGYTMEQMFEVVSDVGSYNKFLPWCKKSIVLKETPGNLKADLIIGFPPINESYTSNVTLVKPHLVKAECSDGRLFHHMLTLWRFSPGLKREQQSCVVDFQITFEFRSAIHSHLSNLFFDQVARQMEGAFIKEVGRRNGPATMQPRKLLINNHTLKT
Summary
Description
Required for the function of coenzyme Q in the respiratory chain. May serve as a chaperone or may be involved in the transport of Q6 from its site of synthesis to the catalytic sites of the respiratory complexes (Probable).
Required for the function of coenzyme Q in the respiratory chain. May serve as a chaperone or may be involved in the transport of Q6 from its site of synthesis to the catalytic sites of the respiratory complexes (By similarity).
Required for the function of coenzyme Q in the respiratory chain. May serve as a chaperone or may be involved in the transport of Q6 from its site of synthesis to the catalytic sites of the respiratory complexes (By similarity).
Subunit
Interacts with coenzyme Q.
Similarity
Belongs to the COQ10 family.
Keywords
Alternative splicing
Complete proteome
Membrane
Mitochondrion
Mitochondrion inner membrane
Polymorphism
Reference proteome
Transit peptide
Feature
chain Coenzyme Q-binding protein COQ10 homolog A, mitochondrial
splice variant In isoform 3.
sequence variant In dbSNP:rs11543258.
splice variant In isoform 3.
sequence variant In dbSNP:rs11543258.
Uniprot
A0A0L7LF54
A0A2A4JRS7
A0A2H1WNY0
A0A194QMJ5
A0A1C9EGN9
A0A212EQ14
+ More
A0A194PE91 B0WT35 A0A182GLV8 Q17FA5 A0A1S4F514 A0A182Q9K6 A0A084WN32 A0A023ELN3 A0A182ILW3 A0A182MAX5 A0A182JR02 A0A182UQU9 A0A182X3W4 A0A182KR54 Q7QJB2 A0A182I358 A0A182XYR1 A0A182UFD8 A0A182NI07 A0A182R7C6 A0A182PQK2 A0A182WJD0 A0A2M3ZJM4 A0A2M4BZK4 A0A2M4BZX5 A0A2P8YGT9 A0A182FGV5 A0A2M4AUI3 A0A2M4AUI7 A0A0M8ZSL3 A0A2M4CPL6 W5JVB5 A0A088APU3 A0A2A3EI74 A0A195EYY0 A0A0P5I671 A0A195DFC5 A0A0P5SJ14 M7BG08 A0A0P5EBX7 F4W5S7 A0A151I2Y3 A0A2P0XJ08 A0A158P3B6 K7F5S6 A0A151X331 A0A0L7R7S4 A0A0P5GYN5 A0A3Q7W456 G3VEY6 H3BI65 E9IPJ8 A0A0P9AHJ2 G1KGQ9 B3MJ38 A0A067RHE7 A0A3Q7X0B7 G3TE04 A0A384BJ21 A0A1W4V1J6 F7EQA5 A0A2K6FGW5 A0A2P4S957 B4LK00 A0A3Q7SPD1 A0A026VTU7 A0A0G2JYY6 G7N7C3 K7BCP7 A0A232EQV1 A0A0Q9W5D6 Q96MF6-3 G3R133 A0A226NQS4 L5KWT1 A0A2K6S2D5 J9P3S8 A0A3L7IHG9 A0A1L8HHW7 Q6DFA6 A0A226MED8 A0A091V9T8 R4UL61 A0A2K6FGW2 G1LAP7 A0A337SIC9 A0A2J8UHU3 F7EC97 A0A2K6K4P0 A0A2K6Q6T6 A0A2K5L2C1
A0A194PE91 B0WT35 A0A182GLV8 Q17FA5 A0A1S4F514 A0A182Q9K6 A0A084WN32 A0A023ELN3 A0A182ILW3 A0A182MAX5 A0A182JR02 A0A182UQU9 A0A182X3W4 A0A182KR54 Q7QJB2 A0A182I358 A0A182XYR1 A0A182UFD8 A0A182NI07 A0A182R7C6 A0A182PQK2 A0A182WJD0 A0A2M3ZJM4 A0A2M4BZK4 A0A2M4BZX5 A0A2P8YGT9 A0A182FGV5 A0A2M4AUI3 A0A2M4AUI7 A0A0M8ZSL3 A0A2M4CPL6 W5JVB5 A0A088APU3 A0A2A3EI74 A0A195EYY0 A0A0P5I671 A0A195DFC5 A0A0P5SJ14 M7BG08 A0A0P5EBX7 F4W5S7 A0A151I2Y3 A0A2P0XJ08 A0A158P3B6 K7F5S6 A0A151X331 A0A0L7R7S4 A0A0P5GYN5 A0A3Q7W456 G3VEY6 H3BI65 E9IPJ8 A0A0P9AHJ2 G1KGQ9 B3MJ38 A0A067RHE7 A0A3Q7X0B7 G3TE04 A0A384BJ21 A0A1W4V1J6 F7EQA5 A0A2K6FGW5 A0A2P4S957 B4LK00 A0A3Q7SPD1 A0A026VTU7 A0A0G2JYY6 G7N7C3 K7BCP7 A0A232EQV1 A0A0Q9W5D6 Q96MF6-3 G3R133 A0A226NQS4 L5KWT1 A0A2K6S2D5 J9P3S8 A0A3L7IHG9 A0A1L8HHW7 Q6DFA6 A0A226MED8 A0A091V9T8 R4UL61 A0A2K6FGW2 G1LAP7 A0A337SIC9 A0A2J8UHU3 F7EC97 A0A2K6K4P0 A0A2K6Q6T6 A0A2K5L2C1
Pubmed
26227816
26354079
22118469
26483478
17510324
24438588
+ More
24945155 20966253 12364791 14747013 17210077 25244985 29403074 20920257 23761445 23624526 21719571 21347285 17381049 21709235 9215903 21282665 17994087 24845553 25243066 24508170 15057822 22002653 16136131 28648823 14702039 15489334 12975309 16230336 22398555 24621616 23258410 16341006 29704459 27762356 20010809 17975172 25362486
24945155 20966253 12364791 14747013 17210077 25244985 29403074 20920257 23761445 23624526 21719571 21347285 17381049 21709235 9215903 21282665 17994087 24845553 25243066 24508170 15057822 22002653 16136131 28648823 14702039 15489334 12975309 16230336 22398555 24621616 23258410 16341006 29704459 27762356 20010809 17975172 25362486
EMBL
JTDY01001386
KOB74014.1
NWSH01000751
PCG74398.1
ODYU01009998
SOQ54783.1
+ More
KQ461198 KPJ06175.1 KX778610 AON96655.1 AGBW02013342 OWR43566.1 KQ459606 KPI91582.1 DS232080 EDS34178.1 JXUM01073165 JXUM01073166 KQ562759 KXJ75180.1 CH477272 EAT45242.1 AXCN02000386 ATLV01024557 KE525352 KFB51626.1 GAPW01003430 JAC10168.1 AXCM01001984 AAAB01008807 EAA04466.4 APCN01000199 GGFM01007958 MBW28709.1 GGFJ01009311 MBW58452.1 GGFJ01009441 MBW58582.1 PYGN01000604 PSN43467.1 GGFK01011119 MBW44440.1 GGFK01011135 MBW44456.1 KQ435922 KOX68679.1 GGFL01002953 MBW67131.1 ADMH02000243 ETN67263.1 KZ288232 PBC31493.1 KQ981905 KYN33336.1 GDIQ01224216 JAK27509.1 KQ980903 KYN11598.1 GDIP01142962 JAL60752.1 KB547351 EMP31033.1 GDIP01145112 JAJ78290.1 GL887695 EGI70403.1 KQ976518 KYM82194.1 KY661329 AVA17416.1 ADTU01007401 AGCU01140800 AGCU01140801 KQ982576 KYQ54660.1 KQ414639 KOC66826.1 GDIQ01256976 JAJ94748.1 AEFK01210264 AFYH01001767 AFYH01001768 GL764539 EFZ17503.1 CH902619 KPU77287.1 EDV38132.1 KK852470 KDR23197.1 GAMT01010187 GAMS01003624 GAMR01002432 GAMQ01005084 GAMP01009817 JAB01674.1 JAB19512.1 JAB31500.1 JAB36767.1 JAB42938.1 PPHD01080082 POI20652.1 CH940648 EDW61654.1 KK107921 EZA47213.1 AABR07073270 CM001263 EHH20858.1 AACZ04012964 GABC01007033 GABF01004182 GABE01003224 NBAG03000092 JAA04305.1 JAA17963.1 JAA41515.1 PNI90180.1 NNAY01002718 OXU20697.1 KRF80125.1 AK057003 AK057014 BC002435 BC026922 BC047444 BC073923 AY358861 CABD030083604 AWGT02002303 OXB70206.1 KB030539 ELK15278.1 AAEX03006926 RAZU01000050 RLQ77557.1 CM004468 OCT95665.1 BC076834 MCFN01001117 OXB53646.1 KK733824 KFR00092.1 KC741055 AGM32879.1 ACTA01138067 ACTA01146066 AANG04003655 NDHI03003457 PNJ44793.1
KQ461198 KPJ06175.1 KX778610 AON96655.1 AGBW02013342 OWR43566.1 KQ459606 KPI91582.1 DS232080 EDS34178.1 JXUM01073165 JXUM01073166 KQ562759 KXJ75180.1 CH477272 EAT45242.1 AXCN02000386 ATLV01024557 KE525352 KFB51626.1 GAPW01003430 JAC10168.1 AXCM01001984 AAAB01008807 EAA04466.4 APCN01000199 GGFM01007958 MBW28709.1 GGFJ01009311 MBW58452.1 GGFJ01009441 MBW58582.1 PYGN01000604 PSN43467.1 GGFK01011119 MBW44440.1 GGFK01011135 MBW44456.1 KQ435922 KOX68679.1 GGFL01002953 MBW67131.1 ADMH02000243 ETN67263.1 KZ288232 PBC31493.1 KQ981905 KYN33336.1 GDIQ01224216 JAK27509.1 KQ980903 KYN11598.1 GDIP01142962 JAL60752.1 KB547351 EMP31033.1 GDIP01145112 JAJ78290.1 GL887695 EGI70403.1 KQ976518 KYM82194.1 KY661329 AVA17416.1 ADTU01007401 AGCU01140800 AGCU01140801 KQ982576 KYQ54660.1 KQ414639 KOC66826.1 GDIQ01256976 JAJ94748.1 AEFK01210264 AFYH01001767 AFYH01001768 GL764539 EFZ17503.1 CH902619 KPU77287.1 EDV38132.1 KK852470 KDR23197.1 GAMT01010187 GAMS01003624 GAMR01002432 GAMQ01005084 GAMP01009817 JAB01674.1 JAB19512.1 JAB31500.1 JAB36767.1 JAB42938.1 PPHD01080082 POI20652.1 CH940648 EDW61654.1 KK107921 EZA47213.1 AABR07073270 CM001263 EHH20858.1 AACZ04012964 GABC01007033 GABF01004182 GABE01003224 NBAG03000092 JAA04305.1 JAA17963.1 JAA41515.1 PNI90180.1 NNAY01002718 OXU20697.1 KRF80125.1 AK057003 AK057014 BC002435 BC026922 BC047444 BC073923 AY358861 CABD030083604 AWGT02002303 OXB70206.1 KB030539 ELK15278.1 AAEX03006926 RAZU01000050 RLQ77557.1 CM004468 OCT95665.1 BC076834 MCFN01001117 OXB53646.1 KK733824 KFR00092.1 KC741055 AGM32879.1 ACTA01138067 ACTA01146066 AANG04003655 NDHI03003457 PNJ44793.1
Proteomes
UP000037510
UP000218220
UP000053240
UP000007151
UP000053268
UP000002320
+ More
UP000069940 UP000249989 UP000008820 UP000075886 UP000030765 UP000075880 UP000075883 UP000075881 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000076408 UP000075902 UP000075884 UP000075900 UP000075885 UP000075920 UP000245037 UP000069272 UP000053105 UP000000673 UP000005203 UP000242457 UP000078541 UP000078492 UP000031443 UP000007755 UP000078540 UP000005205 UP000007267 UP000075809 UP000053825 UP000286642 UP000007648 UP000008672 UP000007801 UP000001646 UP000027135 UP000007646 UP000261680 UP000192221 UP000008225 UP000233160 UP000008792 UP000286640 UP000053097 UP000002494 UP000002277 UP000215335 UP000005640 UP000001519 UP000198419 UP000010552 UP000233220 UP000002254 UP000273346 UP000186698 UP000198323 UP000053605 UP000008912 UP000011712 UP000233180 UP000233200 UP000233060
UP000069940 UP000249989 UP000008820 UP000075886 UP000030765 UP000075880 UP000075883 UP000075881 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000076408 UP000075902 UP000075884 UP000075900 UP000075885 UP000075920 UP000245037 UP000069272 UP000053105 UP000000673 UP000005203 UP000242457 UP000078541 UP000078492 UP000031443 UP000007755 UP000078540 UP000005205 UP000007267 UP000075809 UP000053825 UP000286642 UP000007648 UP000008672 UP000007801 UP000001646 UP000027135 UP000007646 UP000261680 UP000192221 UP000008225 UP000233160 UP000008792 UP000286640 UP000053097 UP000002494 UP000002277 UP000215335 UP000005640 UP000001519 UP000198419 UP000010552 UP000233220 UP000002254 UP000273346 UP000186698 UP000198323 UP000053605 UP000008912 UP000011712 UP000233180 UP000233200 UP000233060
PRIDE
Pfam
PF03364 Polyketide_cyc
Gene 3D
ProteinModelPortal
A0A0L7LF54
A0A2A4JRS7
A0A2H1WNY0
A0A194QMJ5
A0A1C9EGN9
A0A212EQ14
+ More
A0A194PE91 B0WT35 A0A182GLV8 Q17FA5 A0A1S4F514 A0A182Q9K6 A0A084WN32 A0A023ELN3 A0A182ILW3 A0A182MAX5 A0A182JR02 A0A182UQU9 A0A182X3W4 A0A182KR54 Q7QJB2 A0A182I358 A0A182XYR1 A0A182UFD8 A0A182NI07 A0A182R7C6 A0A182PQK2 A0A182WJD0 A0A2M3ZJM4 A0A2M4BZK4 A0A2M4BZX5 A0A2P8YGT9 A0A182FGV5 A0A2M4AUI3 A0A2M4AUI7 A0A0M8ZSL3 A0A2M4CPL6 W5JVB5 A0A088APU3 A0A2A3EI74 A0A195EYY0 A0A0P5I671 A0A195DFC5 A0A0P5SJ14 M7BG08 A0A0P5EBX7 F4W5S7 A0A151I2Y3 A0A2P0XJ08 A0A158P3B6 K7F5S6 A0A151X331 A0A0L7R7S4 A0A0P5GYN5 A0A3Q7W456 G3VEY6 H3BI65 E9IPJ8 A0A0P9AHJ2 G1KGQ9 B3MJ38 A0A067RHE7 A0A3Q7X0B7 G3TE04 A0A384BJ21 A0A1W4V1J6 F7EQA5 A0A2K6FGW5 A0A2P4S957 B4LK00 A0A3Q7SPD1 A0A026VTU7 A0A0G2JYY6 G7N7C3 K7BCP7 A0A232EQV1 A0A0Q9W5D6 Q96MF6-3 G3R133 A0A226NQS4 L5KWT1 A0A2K6S2D5 J9P3S8 A0A3L7IHG9 A0A1L8HHW7 Q6DFA6 A0A226MED8 A0A091V9T8 R4UL61 A0A2K6FGW2 G1LAP7 A0A337SIC9 A0A2J8UHU3 F7EC97 A0A2K6K4P0 A0A2K6Q6T6 A0A2K5L2C1
A0A194PE91 B0WT35 A0A182GLV8 Q17FA5 A0A1S4F514 A0A182Q9K6 A0A084WN32 A0A023ELN3 A0A182ILW3 A0A182MAX5 A0A182JR02 A0A182UQU9 A0A182X3W4 A0A182KR54 Q7QJB2 A0A182I358 A0A182XYR1 A0A182UFD8 A0A182NI07 A0A182R7C6 A0A182PQK2 A0A182WJD0 A0A2M3ZJM4 A0A2M4BZK4 A0A2M4BZX5 A0A2P8YGT9 A0A182FGV5 A0A2M4AUI3 A0A2M4AUI7 A0A0M8ZSL3 A0A2M4CPL6 W5JVB5 A0A088APU3 A0A2A3EI74 A0A195EYY0 A0A0P5I671 A0A195DFC5 A0A0P5SJ14 M7BG08 A0A0P5EBX7 F4W5S7 A0A151I2Y3 A0A2P0XJ08 A0A158P3B6 K7F5S6 A0A151X331 A0A0L7R7S4 A0A0P5GYN5 A0A3Q7W456 G3VEY6 H3BI65 E9IPJ8 A0A0P9AHJ2 G1KGQ9 B3MJ38 A0A067RHE7 A0A3Q7X0B7 G3TE04 A0A384BJ21 A0A1W4V1J6 F7EQA5 A0A2K6FGW5 A0A2P4S957 B4LK00 A0A3Q7SPD1 A0A026VTU7 A0A0G2JYY6 G7N7C3 K7BCP7 A0A232EQV1 A0A0Q9W5D6 Q96MF6-3 G3R133 A0A226NQS4 L5KWT1 A0A2K6S2D5 J9P3S8 A0A3L7IHG9 A0A1L8HHW7 Q6DFA6 A0A226MED8 A0A091V9T8 R4UL61 A0A2K6FGW2 G1LAP7 A0A337SIC9 A0A2J8UHU3 F7EC97 A0A2K6K4P0 A0A2K6Q6T6 A0A2K5L2C1
PDB
1T17
E-value=1.81321e-05,
Score=112
Ontologies
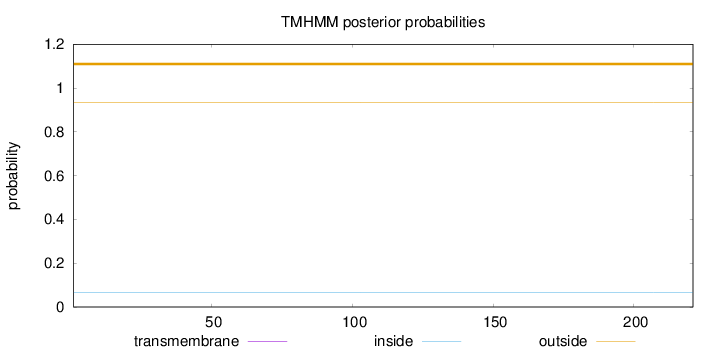
Topology
Subcellular location
Mitochondrion inner membrane
Length:
221
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00141
Exp number, first 60 AAs:
0.000880000000000001
Total prob of N-in:
0.06734
outside
1 - 221
Population Genetic Test Statistics
Pi
292.861071
Theta
217.659136
Tajima's D
1.127078
CLR
0.000446
CSRT
0.687415629218539
Interpretation
Uncertain
