Gene
KWMTBOMO05609
Pre Gene Modal
BGIBMGA006770
Annotation
PREDICTED:_sterile_alpha_and_TIR_motif-containing_protein_1_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.833
Sequence
CDS
ATGGCCTCGTACTCCACATACACGGGAAAACCAAAAATTTTCTTCCCAAAGAAGGTCCGTTCCGAAGAGTCCCTTCTTGCACAAGAAAAGATCCCGAAGAACTCTCGATGGATACCTGAGCTTACGCTCGATGGAAGCAACAGTCTGAAACACAACGGAAGCGCCAAAGACTTGGCATCGGTCATGGAGAATCTAAAGAAGGGCGGTCTGGTCTCCAGACAAAACCAGTCCTTAAATACACAAGTCACTAGTTCGACGCAACAGATGGTCTCCAGTACTACGTCGAGTAGCGCGGCCTCCAGTCAGCGTATGCAAAGTTCTTCCCAAAGACTGCAGCACATGGCAACTTCGGAAATGAAAGCCAGCTCGATGAAATCTGATCTGACTGAACTAAAAAGTTCGATTTCCGAAATGAAGAATCTAAGCAGTACAGCAGCTAAAAACTTTGGGCAAAGATTGCGAAGCTCCATGGAAAACATAGTAGATCAAGATGATATGGGCGAACGCCACCTTCCTGATATACGAGATCTTGGTGAAATAAGCGATTTGGGTAATATGAGTGATATGAGCGAACTCCAAGGGGACGGACCACTGGTGACTTTTCCCGAATCCGATACGCCACCTCCTATGAGTGAAAAACCATCTTTGCTTTCTGGAAACACAGTAGCCGTTGGATCAAGTGCTGCAAATGAATTGAAGTACAGTGCGGCACGGGTCACGTCAGCTAGTTCGACCCGGGTTGTAGCTGACGGTTACAGTGCATCAAGATCAGCCGCGAACAGTGCAAGGATGCGTCGTCTACAGCACGGCGAACTGCAGTATGCTGAAAGAAGTGCGGCTGGAGCATCACATCGACTGCTACAGGCCGAAGGACTTACTGCCGAACAAAACGCCGCGTACCTTCAAGAGCAGCGTTCCTTAAAAGCGGGTGAAATGAGTGCGCAAGAAGCTAATAATATGTCCGCTTCGAGTTCGAGACTGCAAACCGAAGCTTTCAGTGCCGAGAAAAAAGCTATGGCTTCAGCGCAAGCACGACAAACTGTCACATCCAGTGGGATGTTCTCTCACAAGGAACACTCCACTATGGCACAATCGAATATGACAATCTCGAGCAAGAACCTCACAACCAAATCAACGCTTCTCTCCTCACAGATGAGTCAATTACTAAATGGGACCATCAAACCCGGCGACGAAGATCTCACAAATTTAACTTTCGAGGATTTAAACAATCTAAACGCCAATTCAAATCAAAAGGATGTAGATATGGCCATACAGAAATATTCATCGCGAATGAACGCTTTCATTACATCCATAAAAAACAAACAAATGGATATGAAAAATGCATCTATACATTTCAGTAAACTGAATGAAATGTTGAGGAGAGCTTGGGCGGTACCTACATATGGACACGAACTTGGCTATTCACTTTGCAATACATTGCGGACTTCGGGTGGCTTAGACATTCTGATGGCGAATTGTTTGGAGTCTAACAATCCGGAACTGCAGTTCTCATCTGCCAAATTACTTGAGCAATGTCTCACGACAGAAAACAGGGCGCACGTCGTAGAGAATGGTCTCGAAAAGGTCGTAAACGTCGCATGTGTTTGTACGAAGAATGCGAATTCCGTGGACCACTCTCGAGTTGGTACAGGCATTTTGGAGCACCTGTTCAAGCACAGCGAGAGCACCTGCGATGACGTCATCAAATTAGGAGGTCTGGATGCGGTGCTGTTCGAATGCAGAAAAAACGACGTGGAAACTTTGAGACACTGCGCGACGGCTCTAGCGAACTTATCTCTATACGGTGGCGCGGAAAACCAAGAAGCTATGATCAAGAGAAAAGTACCGATGTGGCTGTTTCCTTTGGCTTTTCATAACGACGACAACATTAAATATTACGCGTGCTTAGCTATAGCGGTCCTAGTCGCTAACAAAGAAATCGAAGCCGCAGTATTGAAATCTGGGACATTAGATTTGGTCGAACCGTTCGTTACGTCACACAATCCTTCCGAATTCGCGAGATCGAATCTCGCTCACGCGCACGGTCAAAGCAAAAATTGGCTGAAAAGATTAGTTCCAGTGTTGAGTTCGAAGAGGGAAGAAGCTAGGAATCTGGCGGCGTTCCACTTCTGCATGGAAGCCGGGATCAAGAAGCAACAGGGGAACACGGAGATCTTTAGAGAAATAGGTGCCATAGAATCGCTCAAGAAAGTTGCCAGTTGCCCAAACGCGGTGGCCTCTAAATACGCCGCGCAGGCTCTTCGACTTATCGGGGAGGAAGTTCCACACAAGTTATCACAGCAGGTTCCCCTCTGGTCAATCGAAGACGTAAGAGAATGGGTCAAACAAATAGGGTTTTCGGAATACGCTACAAACTTTTACGAAAGCAGAGTCGACGGTGATTTACTGTTGCAAATCACGGAAGAGAATTTAAAAGAAGACATAGGCTTACAAAATGGGATCAAAAGAAAAAGATTTACCCGCGAACTCCAACAATTAAAGAAAATGGCCGATTACACGTCTCGCGACACAGGGAGTCTGAATGAGTTTTTACAAAGCATTGGTCCGGAATACACTATATACACGTACTCTATGCTCAACGCTGGCGTAGATAAAGAATCTATTCGAGGCCTGAGCGACGAGCAATTGGAAAGCGAATGTCGCATTGGCAACAGCATTCACAGACTAAGGATATTGAATGCCATCAAAGCTTACGAAAACAGTTTACCCAGTAAAGGCGAAGAGAACATGGAGAAGAATTTGGACGTCTTCGTCAGCTATAGAAGGTCAAATGGATCGCAGCTGGCTAGCTTGCTCAAAGTGCACTTACAGTTGAGAGGTTTCACGGTGTTCATCGATGTGGAGAGGCTAGAAGCAGGCAAATTTGACAATAATTTGCTGCAGAGTATCAGACAAGCTAAACACTTCTTGCTTGTTCTCACACCGAACGCCTTGGACAGGTGTAAGAGTGATCATGACAGAAAAGACTGGGTCCATCGGGAAATTGTAGCAGCGTTACAATCGCAGTGTAACATTGTGCCAATTATCGACAACTTCGAATGGCCTAAAGCTGAGGAGTTGCCTGAAGATATGAGAGCCGTGTGCCATTTCAACGGAGTCCGGTGGATTCACGACTACCAAGACGCTTGCGTCGAAAAACTTGAAAGTTTCCTACGTGGAAAGTCTAATCTCGCCAATCGCCTAGAGGGACAGCTCAGAGGTCGCGGGGACGTCCAAACCCCCGGGACCGCTACGATAGGTCGCGGTCCGCCGAACTATCAACGAATGGCGTCTTGCGAGAGCCGCGGTAGTGATAAAGCAGATTGA
Protein
MASYSTYTGKPKIFFPKKVRSEESLLAQEKIPKNSRWIPELTLDGSNSLKHNGSAKDLASVMENLKKGGLVSRQNQSLNTQVTSSTQQMVSSTTSSSAASSQRMQSSSQRLQHMATSEMKASSMKSDLTELKSSISEMKNLSSTAAKNFGQRLRSSMENIVDQDDMGERHLPDIRDLGEISDLGNMSDMSELQGDGPLVTFPESDTPPPMSEKPSLLSGNTVAVGSSAANELKYSAARVTSASSTRVVADGYSASRSAANSARMRRLQHGELQYAERSAAGASHRLLQAEGLTAEQNAAYLQEQRSLKAGEMSAQEANNMSASSSRLQTEAFSAEKKAMASAQARQTVTSSGMFSHKEHSTMAQSNMTISSKNLTTKSTLLSSQMSQLLNGTIKPGDEDLTNLTFEDLNNLNANSNQKDVDMAIQKYSSRMNAFITSIKNKQMDMKNASIHFSKLNEMLRRAWAVPTYGHELGYSLCNTLRTSGGLDILMANCLESNNPELQFSSAKLLEQCLTTENRAHVVENGLEKVVNVACVCTKNANSVDHSRVGTGILEHLFKHSESTCDDVIKLGGLDAVLFECRKNDVETLRHCATALANLSLYGGAENQEAMIKRKVPMWLFPLAFHNDDNIKYYACLAIAVLVANKEIEAAVLKSGTLDLVEPFVTSHNPSEFARSNLAHAHGQSKNWLKRLVPVLSSKREEARNLAAFHFCMEAGIKKQQGNTEIFREIGAIESLKKVASCPNAVASKYAAQALRLIGEEVPHKLSQQVPLWSIEDVREWVKQIGFSEYATNFYESRVDGDLLLQITEENLKEDIGLQNGIKRKRFTRELQQLKKMADYTSRDTGSLNEFLQSIGPEYTIYTYSMLNAGVDKESIRGLSDEQLESECRIGNSIHRLRILNAIKAYENSLPSKGEENMEKNLDVFVSYRRSNGSQLASLLKVHLQLRGFTVFIDVERLEAGKFDNNLLQSIRQAKHFLLVLTPNALDRCKSDHDRKDWVHREIVAALQSQCNIVPIIDNFEWPKAEELPEDMRAVCHFNGVRWIHDYQDACVEKLESFLRGKSNLANRLEGQLRGRGDVQTPGTATIGRGPPNYQRMASCESRGSDKAD
Summary
Uniprot
A0A2A4K0D8
A0A2A4K053
A0A2A4K0H2
A0A2H1WNY4
A0A2A4JYN9
A0A3S2NJB0
+ More
A0A2A4JZN0 A0A1C9EGN4 H9JB75 A0A2A4JYP0 A0A2A4JZA3 A0A212EPZ9 A0A194QMJ8 A0A194PKG4 A0A195AV14 A0A195E8Z1 A0A151IPW8 A0A158NET3 A0A195FN53 A0A2J7RJG7 A0A067QS80 A0A1Y1NIE9 A0A1Y1NE91 A0A154PJ55 A0A3L8DCF7 A0A026W3B1 A0A0C9RAX8 A0A1Y1ND97 A0A1Y1ND63 A0A0C9RK29 A0A1B6C901 A0A0C9QGG6 A0A0C9RKT0 A0A1L8E3C9 A0A1L8E3T8 A0A0J7L3C8 E2B7A2 A0A1L8E3D7 E2A4V6 A0A139WBM7 A0A023F2Z7 A0A139WBJ6 A0A069DY07 A0A2A3ELK1 A0A088AIZ8 F4WEQ2 A0A0V0G4S2 A0A224XIK3 A0A0C9RX31 E0V9G8 A0A0L7RJ69 A0A1I8N513 A0A1I8N519 A0A1I8N514 A0A1I8N520 A0A1I8N516 T1PBZ0 T1PDJ5 A0A1I8N530 A0A0K8SPG2 W8AAV2 A0A0K8SPA2 A0A0K8SPX7 A0A0K8VGS6 A0A1Q3G4X5 A0A1Q3G4Y7 A0A1J1I3H0 A0A034V4J3 A0A0P8ZVC0 A0A0P8XUY8 A0A0Q5UDG0 A0A2M3ZGD8 A0A0P8YAS0 A0A1W4VAC0 A0A0K8VTW8 A0A0P9AL23 A0A0Q5U4T3 A0A0K8UC39 A0A0K8WFQ8 A0A0Q5U331 A0A0Q5U2B8 A0A0Q5U2P8 A0A0Q5UHD8 A0A1W4VM22 A0A1W4VNF5 Q16F20 A0A0J9RR15 A0A0R1DWJ1 A0A0R1DWE8 A0A0Q5UHP6 A0A1W4VMR7 A0A0J9RRD8 A0A0J9RQW5 A0A182QCA4 A8JNN2 A8JNN1 M9PHR5 A0A0R1E184
A0A2A4JZN0 A0A1C9EGN4 H9JB75 A0A2A4JYP0 A0A2A4JZA3 A0A212EPZ9 A0A194QMJ8 A0A194PKG4 A0A195AV14 A0A195E8Z1 A0A151IPW8 A0A158NET3 A0A195FN53 A0A2J7RJG7 A0A067QS80 A0A1Y1NIE9 A0A1Y1NE91 A0A154PJ55 A0A3L8DCF7 A0A026W3B1 A0A0C9RAX8 A0A1Y1ND97 A0A1Y1ND63 A0A0C9RK29 A0A1B6C901 A0A0C9QGG6 A0A0C9RKT0 A0A1L8E3C9 A0A1L8E3T8 A0A0J7L3C8 E2B7A2 A0A1L8E3D7 E2A4V6 A0A139WBM7 A0A023F2Z7 A0A139WBJ6 A0A069DY07 A0A2A3ELK1 A0A088AIZ8 F4WEQ2 A0A0V0G4S2 A0A224XIK3 A0A0C9RX31 E0V9G8 A0A0L7RJ69 A0A1I8N513 A0A1I8N519 A0A1I8N514 A0A1I8N520 A0A1I8N516 T1PBZ0 T1PDJ5 A0A1I8N530 A0A0K8SPG2 W8AAV2 A0A0K8SPA2 A0A0K8SPX7 A0A0K8VGS6 A0A1Q3G4X5 A0A1Q3G4Y7 A0A1J1I3H0 A0A034V4J3 A0A0P8ZVC0 A0A0P8XUY8 A0A0Q5UDG0 A0A2M3ZGD8 A0A0P8YAS0 A0A1W4VAC0 A0A0K8VTW8 A0A0P9AL23 A0A0Q5U4T3 A0A0K8UC39 A0A0K8WFQ8 A0A0Q5U331 A0A0Q5U2B8 A0A0Q5U2P8 A0A0Q5UHD8 A0A1W4VM22 A0A1W4VNF5 Q16F20 A0A0J9RR15 A0A0R1DWJ1 A0A0R1DWE8 A0A0Q5UHP6 A0A1W4VMR7 A0A0J9RRD8 A0A0J9RQW5 A0A182QCA4 A8JNN2 A8JNN1 M9PHR5 A0A0R1E184
Pubmed
EMBL
NWSH01000351
PCG77140.1
PCG77143.1
PCG77142.1
ODYU01009998
SOQ54780.1
+ More
PCG77135.1 RSAL01000073 RVE49008.1 PCG77138.1 KX778610 AON96652.1 BABH01008720 PCG77141.1 PCG77139.1 AGBW02013342 OWR43570.1 KQ461198 KPJ06180.1 KQ459606 KPI91585.1 KQ976736 KYM76083.1 KQ979479 KYN21297.1 KQ976806 KYN08197.1 ADTU01013666 ADTU01013667 KQ981490 KYN41319.1 NEVH01002992 PNF40972.1 KK853689 KDQ97276.1 GEZM01005824 JAV95886.1 GEZM01005823 JAV95888.1 KQ434936 KZC11875.1 QOIP01000010 RLU17822.1 KK107455 EZA50513.1 GBYB01013644 JAG83411.1 GEZM01005826 JAV95882.1 GEZM01005825 JAV95884.1 GBYB01013647 JAG83414.1 GEDC01027322 JAS09976.1 GBYB01013643 JAG83410.1 GBYB01013912 JAG83679.1 GFDF01000841 JAV13243.1 GFDF01000842 JAV13242.1 LBMM01000947 KMQ97161.1 GL446149 EFN88463.1 GFDF01000847 JAV13237.1 GL436754 EFN71525.1 KQ971372 KYB25339.1 GBBI01002987 JAC15725.1 KYB25338.1 GBGD01000282 JAC88607.1 KZ288226 PBC31901.1 GL888103 EGI67415.1 GECL01003089 JAP03035.1 GFTR01008463 JAW07963.1 GBYB01013645 JAG83412.1 DS234993 EEB10024.1 KQ414582 KOC70905.1 KA646204 AFP60833.1 KA646205 AFP60834.1 GBRD01010648 GBRD01010646 JAG55176.1 GAMC01021615 JAB84940.1 GBRD01010647 GBRD01006414 JAG55177.1 GBRD01010650 GBRD01010649 JAG55174.1 GDHF01014247 JAI38067.1 GFDL01000183 JAV34862.1 GFDL01000180 JAV34865.1 CVRI01000035 CRK92905.1 GAKP01022292 JAC36660.1 CH902618 KPU78536.1 KPU78541.1 CH954178 KQS43265.1 GGFM01006769 MBW27520.1 KPU78537.1 GDHF01010016 JAI42298.1 KPU78539.1 KQS43260.1 GDHF01028399 GDHF01005600 JAI23915.1 JAI46714.1 GDHF01002313 JAI50001.1 KQS43259.1 KQS43263.1 KQS43262.1 KQS43264.1 CH478527 EAT32828.1 CM002912 KMY98263.1 CM000159 KRK01421.1 KRK01420.1 KQS43266.1 KMY98267.1 KMY98261.1 AXCN02000924 AE014296 ABW08491.1 ABW08490.1 AGB94251.1 KRK01425.1
PCG77135.1 RSAL01000073 RVE49008.1 PCG77138.1 KX778610 AON96652.1 BABH01008720 PCG77141.1 PCG77139.1 AGBW02013342 OWR43570.1 KQ461198 KPJ06180.1 KQ459606 KPI91585.1 KQ976736 KYM76083.1 KQ979479 KYN21297.1 KQ976806 KYN08197.1 ADTU01013666 ADTU01013667 KQ981490 KYN41319.1 NEVH01002992 PNF40972.1 KK853689 KDQ97276.1 GEZM01005824 JAV95886.1 GEZM01005823 JAV95888.1 KQ434936 KZC11875.1 QOIP01000010 RLU17822.1 KK107455 EZA50513.1 GBYB01013644 JAG83411.1 GEZM01005826 JAV95882.1 GEZM01005825 JAV95884.1 GBYB01013647 JAG83414.1 GEDC01027322 JAS09976.1 GBYB01013643 JAG83410.1 GBYB01013912 JAG83679.1 GFDF01000841 JAV13243.1 GFDF01000842 JAV13242.1 LBMM01000947 KMQ97161.1 GL446149 EFN88463.1 GFDF01000847 JAV13237.1 GL436754 EFN71525.1 KQ971372 KYB25339.1 GBBI01002987 JAC15725.1 KYB25338.1 GBGD01000282 JAC88607.1 KZ288226 PBC31901.1 GL888103 EGI67415.1 GECL01003089 JAP03035.1 GFTR01008463 JAW07963.1 GBYB01013645 JAG83412.1 DS234993 EEB10024.1 KQ414582 KOC70905.1 KA646204 AFP60833.1 KA646205 AFP60834.1 GBRD01010648 GBRD01010646 JAG55176.1 GAMC01021615 JAB84940.1 GBRD01010647 GBRD01006414 JAG55177.1 GBRD01010650 GBRD01010649 JAG55174.1 GDHF01014247 JAI38067.1 GFDL01000183 JAV34862.1 GFDL01000180 JAV34865.1 CVRI01000035 CRK92905.1 GAKP01022292 JAC36660.1 CH902618 KPU78536.1 KPU78541.1 CH954178 KQS43265.1 GGFM01006769 MBW27520.1 KPU78537.1 GDHF01010016 JAI42298.1 KPU78539.1 KQS43260.1 GDHF01028399 GDHF01005600 JAI23915.1 JAI46714.1 GDHF01002313 JAI50001.1 KQS43259.1 KQS43263.1 KQS43262.1 KQS43264.1 CH478527 EAT32828.1 CM002912 KMY98263.1 CM000159 KRK01421.1 KRK01420.1 KQS43266.1 KMY98267.1 KMY98261.1 AXCN02000924 AE014296 ABW08491.1 ABW08490.1 AGB94251.1 KRK01425.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000007151
UP000053240
UP000053268
+ More
UP000078540 UP000078492 UP000078542 UP000005205 UP000078541 UP000235965 UP000027135 UP000076502 UP000279307 UP000053097 UP000036403 UP000008237 UP000000311 UP000007266 UP000242457 UP000005203 UP000007755 UP000009046 UP000053825 UP000095301 UP000183832 UP000007801 UP000008711 UP000192221 UP000008820 UP000002282 UP000075886 UP000000803
UP000078540 UP000078492 UP000078542 UP000005205 UP000078541 UP000235965 UP000027135 UP000076502 UP000279307 UP000053097 UP000036403 UP000008237 UP000000311 UP000007266 UP000242457 UP000005203 UP000007755 UP000009046 UP000053825 UP000095301 UP000183832 UP000007801 UP000008711 UP000192221 UP000008820 UP000002282 UP000075886 UP000000803
Interpro
IPR039184
SARM1
+ More
IPR000157 TIR_dom
IPR016024 ARM-type_fold
IPR013761 SAM/pointed_sf
IPR035897 Toll_tir_struct_dom_sf
IPR011989 ARM-like
IPR001660 SAM
IPR031827 DUF4746
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR011993 PH-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR001849 PH_domain
IPR019775 WD40_repeat_CS
IPR000157 TIR_dom
IPR016024 ARM-type_fold
IPR013761 SAM/pointed_sf
IPR035897 Toll_tir_struct_dom_sf
IPR011989 ARM-like
IPR001660 SAM
IPR031827 DUF4746
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR011993 PH-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR001849 PH_domain
IPR019775 WD40_repeat_CS
ProteinModelPortal
A0A2A4K0D8
A0A2A4K053
A0A2A4K0H2
A0A2H1WNY4
A0A2A4JYN9
A0A3S2NJB0
+ More
A0A2A4JZN0 A0A1C9EGN4 H9JB75 A0A2A4JYP0 A0A2A4JZA3 A0A212EPZ9 A0A194QMJ8 A0A194PKG4 A0A195AV14 A0A195E8Z1 A0A151IPW8 A0A158NET3 A0A195FN53 A0A2J7RJG7 A0A067QS80 A0A1Y1NIE9 A0A1Y1NE91 A0A154PJ55 A0A3L8DCF7 A0A026W3B1 A0A0C9RAX8 A0A1Y1ND97 A0A1Y1ND63 A0A0C9RK29 A0A1B6C901 A0A0C9QGG6 A0A0C9RKT0 A0A1L8E3C9 A0A1L8E3T8 A0A0J7L3C8 E2B7A2 A0A1L8E3D7 E2A4V6 A0A139WBM7 A0A023F2Z7 A0A139WBJ6 A0A069DY07 A0A2A3ELK1 A0A088AIZ8 F4WEQ2 A0A0V0G4S2 A0A224XIK3 A0A0C9RX31 E0V9G8 A0A0L7RJ69 A0A1I8N513 A0A1I8N519 A0A1I8N514 A0A1I8N520 A0A1I8N516 T1PBZ0 T1PDJ5 A0A1I8N530 A0A0K8SPG2 W8AAV2 A0A0K8SPA2 A0A0K8SPX7 A0A0K8VGS6 A0A1Q3G4X5 A0A1Q3G4Y7 A0A1J1I3H0 A0A034V4J3 A0A0P8ZVC0 A0A0P8XUY8 A0A0Q5UDG0 A0A2M3ZGD8 A0A0P8YAS0 A0A1W4VAC0 A0A0K8VTW8 A0A0P9AL23 A0A0Q5U4T3 A0A0K8UC39 A0A0K8WFQ8 A0A0Q5U331 A0A0Q5U2B8 A0A0Q5U2P8 A0A0Q5UHD8 A0A1W4VM22 A0A1W4VNF5 Q16F20 A0A0J9RR15 A0A0R1DWJ1 A0A0R1DWE8 A0A0Q5UHP6 A0A1W4VMR7 A0A0J9RRD8 A0A0J9RQW5 A0A182QCA4 A8JNN2 A8JNN1 M9PHR5 A0A0R1E184
A0A2A4JZN0 A0A1C9EGN4 H9JB75 A0A2A4JYP0 A0A2A4JZA3 A0A212EPZ9 A0A194QMJ8 A0A194PKG4 A0A195AV14 A0A195E8Z1 A0A151IPW8 A0A158NET3 A0A195FN53 A0A2J7RJG7 A0A067QS80 A0A1Y1NIE9 A0A1Y1NE91 A0A154PJ55 A0A3L8DCF7 A0A026W3B1 A0A0C9RAX8 A0A1Y1ND97 A0A1Y1ND63 A0A0C9RK29 A0A1B6C901 A0A0C9QGG6 A0A0C9RKT0 A0A1L8E3C9 A0A1L8E3T8 A0A0J7L3C8 E2B7A2 A0A1L8E3D7 E2A4V6 A0A139WBM7 A0A023F2Z7 A0A139WBJ6 A0A069DY07 A0A2A3ELK1 A0A088AIZ8 F4WEQ2 A0A0V0G4S2 A0A224XIK3 A0A0C9RX31 E0V9G8 A0A0L7RJ69 A0A1I8N513 A0A1I8N519 A0A1I8N514 A0A1I8N520 A0A1I8N516 T1PBZ0 T1PDJ5 A0A1I8N530 A0A0K8SPG2 W8AAV2 A0A0K8SPA2 A0A0K8SPX7 A0A0K8VGS6 A0A1Q3G4X5 A0A1Q3G4Y7 A0A1J1I3H0 A0A034V4J3 A0A0P8ZVC0 A0A0P8XUY8 A0A0Q5UDG0 A0A2M3ZGD8 A0A0P8YAS0 A0A1W4VAC0 A0A0K8VTW8 A0A0P9AL23 A0A0Q5U4T3 A0A0K8UC39 A0A0K8WFQ8 A0A0Q5U331 A0A0Q5U2B8 A0A0Q5U2P8 A0A0Q5UHD8 A0A1W4VM22 A0A1W4VNF5 Q16F20 A0A0J9RR15 A0A0R1DWJ1 A0A0R1DWE8 A0A0Q5UHP6 A0A1W4VMR7 A0A0J9RRD8 A0A0J9RQW5 A0A182QCA4 A8JNN2 A8JNN1 M9PHR5 A0A0R1E184
PDB
3TAD
E-value=0.00073032,
Score=106
Ontologies
GO
PANTHER
Topology
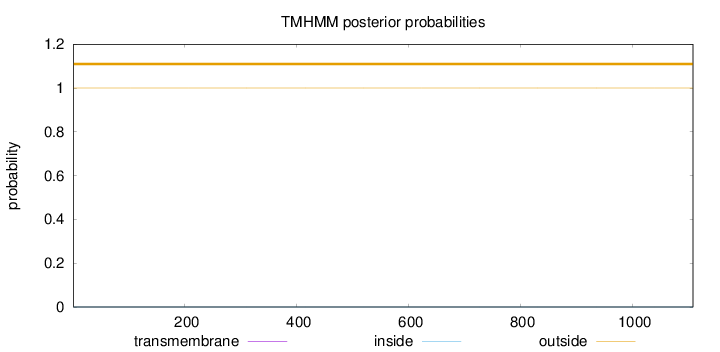
Length:
1108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1108
Population Genetic Test Statistics
Pi
294.664958
Theta
20.013227
Tajima's D
1.525981
CLR
0.370892
CSRT
0.794060296985151
Interpretation
Uncertain
