Gene
KWMTBOMO05607
Pre Gene Modal
BGIBMGA006569
Annotation
PREDICTED:_putative_fatty_acyl-CoA_reductase_CG5065_[Bombyx_mori]
Full name
Fatty acyl-CoA reductase
Location in the cell
PlasmaMembrane Reliability : 2.057
Sequence
CDS
ATGACGTCACAAATAAACGAATGGTATAAAGGCCGGAAAGTCCTTATCACCGGTTCCTCCGGGCTGATGGGCAAGGTCTTAATAGAGAAATTACTGTATAGCGTTCCGGATATAGGATGCGTGTACGCGCTGGTGCGGTCCAAGCGAGGAAAGAGCCCGGAATCACGCATCGAAGACATGTGGAAATTGCCTCTTTTTACGAGAATAAGAGAAGAAAAGCCTCACGTGATGAAGAAGCTTGTAGCCGTCCCTGGCGACATAATCTGCGACAAACTAGGCATCGAAAAAACTAAACTAGAACAAATTTATAATGAGGTTTCGGTAGTTTTCCATTTCGCTGCAAGCTTACGTTTAGAAGCGCCATTGAAGGAAGGCTTGGAAATGAACACAAAAGGCACGATGCGTGTTTTGGAATTATCCAAGAACATAAAAAATTTGGTCGCTTTTGTACATCTGTCGACGGCTTTCTGTTATCCAGATTATGAGCGATTGGCCGAAAAGGTCCACGATCCCCCTACTGATCCCGAAAACGTACTTCAAGCGGCGAGTTGGCTCACCGAACAGCAGTTAAACATTCTCGCTCCTTCAATATACAAGAAGCACCCCAACTCTTACACGTATTCCAAAAGGCTGGCTGAAGCTCTTGTGAAAGAGAGCTACCCTCAACTACCGGCAGTTGTAGTTAGACCTAGTATAGTGACACCCGCATACAAAGAGCCCATAGCAGGTTGGGTTGATAATCTAAACGGCCCCATTGGTTTGATGGTAGGCGCCGGGAAGGGCGTCATCCGCTCCATGCACTGCTACGGTCATTACCACGCCGAGGTCATACCGATAGACATCGCTATTAACGCCATTATAGTTATAAGCTACAAGATTGGAAATCAGTCTGAAAGACCTCCAGAAATACCGACATTCAACCTCACTACAGGAGACGACAGGAACACAACATGGAAGGAAGTTCTGGATATCGGTAAAGCGACCGTTCGAAAGTATCCCTTCGAAGGTCCCCTCTGGTATCCGAACGGCAACATCAGACATAATAAGTTCATACATGACTTGTGTGTGTTCTTCTACCATCTAGTTCCGGCGTATTTTATTGATTTTCTGATGTTTTTGTTCATGCAGAAACGATTTATGGTTCGGATTCAAAATAGGATTTCAGTAGGTCTAGAGGTGCTACAATATTTCACGACGCGTGAATGGTGGTTCGATACAGATAATTTCAAAGCTTTAGCTACAATGTTGAATCCTGTTGATAAGGAAGTTTTTCCTATGGACCTAAGCGTCATTGAAGACGAGCCTTACATAGAGAGCTGTATGATCGGGGGCAAGCTGTATTGCATGAAAGAGAAGCTTGAGAATTTGCCTAAAGCAAGACTCCAGAATAATATTCTCTACGTCTTGGATAAAATAACAACAGTCTTTTTCTACTTGATCCTGCTGTACTGGGTAGTACTGTATTGCGCACCAGTCCGCGAAGTATTATCTTATGGCGGTCCATTTGTTCGATACCTACCTTTGGTGGGTAAAGCGGTTTTTAAAAATGATCTTTAA
Protein
MTSQINEWYKGRKVLITGSSGLMGKVLIEKLLYSVPDIGCVYALVRSKRGKSPESRIEDMWKLPLFTRIREEKPHVMKKLVAVPGDIICDKLGIEKTKLEQIYNEVSVVFHFAASLRLEAPLKEGLEMNTKGTMRVLELSKNIKNLVAFVHLSTAFCYPDYERLAEKVHDPPTDPENVLQAASWLTEQQLNILAPSIYKKHPNSYTYSKRLAEALVKESYPQLPAVVVRPSIVTPAYKEPIAGWVDNLNGPIGLMVGAGKGVIRSMHCYGHYHAEVIPIDIAINAIIVISYKIGNQSERPPEIPTFNLTTGDDRNTTWKEVLDIGKATVRKYPFEGPLWYPNGNIRHNKFIHDLCVFFYHLVPAYFIDFLMFLFMQKRFMVRIQNRISVGLEVLQYFTTREWWFDTDNFKALATMLNPVDKEVFPMDLSVIEDEPYIESCMIGGKLYCMKEKLENLPKARLQNNILYVLDKITTVFFYLILLYWVVLYCAPVREVLSYGGPFVRYLPLVGKAVFKNDL
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Feature
chain Fatty acyl-CoA reductase
Uniprot
H9JAM4
A0A1V0M881
A0A291P0Y2
A0A291P0Q9
A0A2A4K043
A0A2H1VDF8
+ More
U5KG45 A0A068FMK7 A0A1C9EGN8 A0A0F6Q2P8 A0A212EPZ8 I4DM82 A0A194QKX4 A0A194PEZ0 A0A088APG8 A0A3S2NWX2 D9MX50 A0A194QW79 A0A0L7QMT0 A0A0P0DZ32 A0A2H1VBF1 A0A0C9RGI8 A0A2W1BU73 A0A195CPE7 A0A151XJ37 A0A194PRU4 A0A0F6Q2Z9 A0A291P0P8 A0A0F6Q4N3 A0A195FMJ1 A0A0M9A857 A0A1V0M879 A0A2A4JXJ6 A0A182VXI1 U5EPY7 E9ID65 A0A1S4G1B4 A0A182REU5 A0A182V6X2 A0A182I9C8 W5JHR7 A0A182JIX2 Q16GQ4 A0A195EP06 A0A182WVP9 A0A182LXW7 F4WVE2 Q7QAN3 A0A1B0CRT6 A0A0C9QUP1 A0A182KXM4 A0A182FRU7 A0A182QQN9 A0A182NFQ6 A0A1Q3G5B4 A0A182K6D3 A0A1W4XSR6 A0A1W4XHK8 A0A182P917 A0A1L8DYA8 A0A026WTB2 A0A182YI99 A0A084WSE9 A0A067RIF0 A0A1B6C0C4 E2AYW1 A0A195BV13 A0A182SVN7 A0A1B6J616 A0A212FLN3 A0A1Y1KTF8 A0A1B6FF15 A0A1A9UZB6 U4UHU2 A0A034W6Z8 N6TW75 A0A0P8ZQ48 W8BBB7 E0W0P6 B3MC49 B4KQR5 A0A2A3EMS1 A0A0R1DU59 A0A0K8WH42 A0A0K8UGQ7 A0A0Q5W393 B4J9N1 A0A336KQE3 D6WD88 B4PBX4 A0A0Q5VPH3 A0A0J9RKV0 A0A1B6DPR8 A0A0R3NKW5 A0A0J9RL31 A0A0A1XER0 Q8MMC6 Q0E8W1
U5KG45 A0A068FMK7 A0A1C9EGN8 A0A0F6Q2P8 A0A212EPZ8 I4DM82 A0A194QKX4 A0A194PEZ0 A0A088APG8 A0A3S2NWX2 D9MX50 A0A194QW79 A0A0L7QMT0 A0A0P0DZ32 A0A2H1VBF1 A0A0C9RGI8 A0A2W1BU73 A0A195CPE7 A0A151XJ37 A0A194PRU4 A0A0F6Q2Z9 A0A291P0P8 A0A0F6Q4N3 A0A195FMJ1 A0A0M9A857 A0A1V0M879 A0A2A4JXJ6 A0A182VXI1 U5EPY7 E9ID65 A0A1S4G1B4 A0A182REU5 A0A182V6X2 A0A182I9C8 W5JHR7 A0A182JIX2 Q16GQ4 A0A195EP06 A0A182WVP9 A0A182LXW7 F4WVE2 Q7QAN3 A0A1B0CRT6 A0A0C9QUP1 A0A182KXM4 A0A182FRU7 A0A182QQN9 A0A182NFQ6 A0A1Q3G5B4 A0A182K6D3 A0A1W4XSR6 A0A1W4XHK8 A0A182P917 A0A1L8DYA8 A0A026WTB2 A0A182YI99 A0A084WSE9 A0A067RIF0 A0A1B6C0C4 E2AYW1 A0A195BV13 A0A182SVN7 A0A1B6J616 A0A212FLN3 A0A1Y1KTF8 A0A1B6FF15 A0A1A9UZB6 U4UHU2 A0A034W6Z8 N6TW75 A0A0P8ZQ48 W8BBB7 E0W0P6 B3MC49 B4KQR5 A0A2A3EMS1 A0A0R1DU59 A0A0K8WH42 A0A0K8UGQ7 A0A0Q5W393 B4J9N1 A0A336KQE3 D6WD88 B4PBX4 A0A0Q5VPH3 A0A0J9RKV0 A0A1B6DPR8 A0A0R3NKW5 A0A0J9RL31 A0A0A1XER0 Q8MMC6 Q0E8W1
EC Number
1.2.1.84
Pubmed
19121390
28349297
28986331
24053512
26385554
22118469
+ More
22651552 26354079 20542116 26445454 28756777 21282665 20920257 23761445 17510324 21719571 12364791 14747013 17210077 20966253 24508170 30249741 25244985 24438588 24845553 20798317 28004739 23537049 25348373 17994087 24495485 20566863 17550304 18362917 19820115 22936249 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
22651552 26354079 20542116 26445454 28756777 21282665 20920257 23761445 17510324 21719571 12364791 14747013 17210077 20966253 24508170 30249741 25244985 24438588 24845553 20798317 28004739 23537049 25348373 17994087 24495485 20566863 17550304 18362917 19820115 22936249 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01008715
BABH01008716
KU755486
ARD71196.1
MF687593
ATJ44519.1
+ More
MF687538 ATJ44464.1 NWSH01000351 PCG77133.1 ODYU01001961 SOQ38879.1 JX989160 AGR49319.1 KJ622063 AID66651.1 KX778610 AON96651.1 KP237928 AKD01781.1 AGBW02013342 OWR43572.1 AK402400 BAM19022.1 KQ461198 KPJ06182.1 KQ459606 KPI91588.1 RSAL01000039 RVE51027.1 HM536634 ADI87410.1 KQ461108 KPJ09220.1 KQ414868 KOC59957.1 KT261701 ALJ30241.1 ODYU01001651 SOQ38173.1 GBYB01007475 JAG77242.1 KZ149961 PZC76330.1 KQ977444 KYN02613.1 KQ982080 KYQ60351.1 KQ459594 KPI96166.1 KP237917 AKD01770.1 MF687546 ATJ44472.1 KP237930 AKD01783.1 KQ981490 KYN41164.1 KQ435728 KOX77819.1 KU755482 ARD71192.1 NWSH01000478 PCG76112.1 GANO01003516 JAB56355.1 GL762454 EFZ21418.1 APCN01003255 ADMH02001477 ETN62440.1 CH478247 EAT33424.1 KQ978625 KYN29624.1 AXCM01000910 GL888387 EGI61763.1 AAAB01008888 EAA08761.5 AJWK01025254 AJWK01025255 AJWK01025256 GBYB01007474 JAG77241.1 AXCN02000243 GFDL01000089 JAV34956.1 GFDF01002656 JAV11428.1 KK107109 QOIP01000008 EZA59252.1 RLU19596.1 ATLV01026503 ATLV01026504 KE525415 KFB53143.1 KK852458 KDR23567.1 GEDC01030337 JAS06961.1 GL444010 EFN61377.1 KQ976401 KYM92422.1 GECU01013104 JAS94602.1 AGBW02007730 OWR54654.1 GEZM01074076 JAV64682.1 GECZ01020981 JAS48788.1 KB632169 ERL89445.1 GAKP01009042 JAC49910.1 APGK01049539 KB741156 ENN73525.1 CH902619 KPU76653.1 GAMC01010608 JAB95947.1 DS235862 EEB19202.1 EDV37236.2 CH933808 EDW09264.2 KZ288219 PBC32341.1 CM000158 KRK00613.1 GDHF01001846 JAI50468.1 GDHF01026452 JAI25862.1 CH954179 KQS63160.1 CH916367 EDW02538.1 UFQS01000822 UFQT01000822 SSX07044.1 SSX27387.1 KQ971321 EEZ99525.1 EDW92628.2 KQS63158.1 CM002911 KMY96476.1 GEDC01009624 JAS27674.1 CM000071 KRT01617.1 KMY96477.1 GBXI01004852 JAD09440.1 AE013599 BT133449 AAM70799.1 AFH89822.1 AAF47295.2
MF687538 ATJ44464.1 NWSH01000351 PCG77133.1 ODYU01001961 SOQ38879.1 JX989160 AGR49319.1 KJ622063 AID66651.1 KX778610 AON96651.1 KP237928 AKD01781.1 AGBW02013342 OWR43572.1 AK402400 BAM19022.1 KQ461198 KPJ06182.1 KQ459606 KPI91588.1 RSAL01000039 RVE51027.1 HM536634 ADI87410.1 KQ461108 KPJ09220.1 KQ414868 KOC59957.1 KT261701 ALJ30241.1 ODYU01001651 SOQ38173.1 GBYB01007475 JAG77242.1 KZ149961 PZC76330.1 KQ977444 KYN02613.1 KQ982080 KYQ60351.1 KQ459594 KPI96166.1 KP237917 AKD01770.1 MF687546 ATJ44472.1 KP237930 AKD01783.1 KQ981490 KYN41164.1 KQ435728 KOX77819.1 KU755482 ARD71192.1 NWSH01000478 PCG76112.1 GANO01003516 JAB56355.1 GL762454 EFZ21418.1 APCN01003255 ADMH02001477 ETN62440.1 CH478247 EAT33424.1 KQ978625 KYN29624.1 AXCM01000910 GL888387 EGI61763.1 AAAB01008888 EAA08761.5 AJWK01025254 AJWK01025255 AJWK01025256 GBYB01007474 JAG77241.1 AXCN02000243 GFDL01000089 JAV34956.1 GFDF01002656 JAV11428.1 KK107109 QOIP01000008 EZA59252.1 RLU19596.1 ATLV01026503 ATLV01026504 KE525415 KFB53143.1 KK852458 KDR23567.1 GEDC01030337 JAS06961.1 GL444010 EFN61377.1 KQ976401 KYM92422.1 GECU01013104 JAS94602.1 AGBW02007730 OWR54654.1 GEZM01074076 JAV64682.1 GECZ01020981 JAS48788.1 KB632169 ERL89445.1 GAKP01009042 JAC49910.1 APGK01049539 KB741156 ENN73525.1 CH902619 KPU76653.1 GAMC01010608 JAB95947.1 DS235862 EEB19202.1 EDV37236.2 CH933808 EDW09264.2 KZ288219 PBC32341.1 CM000158 KRK00613.1 GDHF01001846 JAI50468.1 GDHF01026452 JAI25862.1 CH954179 KQS63160.1 CH916367 EDW02538.1 UFQS01000822 UFQT01000822 SSX07044.1 SSX27387.1 KQ971321 EEZ99525.1 EDW92628.2 KQS63158.1 CM002911 KMY96476.1 GEDC01009624 JAS27674.1 CM000071 KRT01617.1 KMY96477.1 GBXI01004852 JAD09440.1 AE013599 BT133449 AAM70799.1 AFH89822.1 AAF47295.2
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000005203
+ More
UP000283053 UP000053825 UP000078542 UP000075809 UP000078541 UP000053105 UP000075920 UP000075900 UP000075903 UP000075840 UP000000673 UP000075880 UP000008820 UP000078492 UP000076407 UP000075883 UP000007755 UP000007062 UP000092461 UP000075882 UP000069272 UP000075886 UP000075884 UP000075881 UP000192223 UP000075885 UP000053097 UP000279307 UP000076408 UP000030765 UP000027135 UP000000311 UP000078540 UP000075901 UP000078200 UP000030742 UP000019118 UP000007801 UP000009046 UP000009192 UP000242457 UP000002282 UP000008711 UP000001070 UP000007266 UP000001819 UP000000803
UP000283053 UP000053825 UP000078542 UP000075809 UP000078541 UP000053105 UP000075920 UP000075900 UP000075903 UP000075840 UP000000673 UP000075880 UP000008820 UP000078492 UP000076407 UP000075883 UP000007755 UP000007062 UP000092461 UP000075882 UP000069272 UP000075886 UP000075884 UP000075881 UP000192223 UP000075885 UP000053097 UP000279307 UP000076408 UP000030765 UP000027135 UP000000311 UP000078540 UP000075901 UP000078200 UP000030742 UP000019118 UP000007801 UP000009046 UP000009192 UP000242457 UP000002282 UP000008711 UP000001070 UP000007266 UP000001819 UP000000803
Interpro
SUPFAM
SSF51735
SSF51735
CDD
ProteinModelPortal
H9JAM4
A0A1V0M881
A0A291P0Y2
A0A291P0Q9
A0A2A4K043
A0A2H1VDF8
+ More
U5KG45 A0A068FMK7 A0A1C9EGN8 A0A0F6Q2P8 A0A212EPZ8 I4DM82 A0A194QKX4 A0A194PEZ0 A0A088APG8 A0A3S2NWX2 D9MX50 A0A194QW79 A0A0L7QMT0 A0A0P0DZ32 A0A2H1VBF1 A0A0C9RGI8 A0A2W1BU73 A0A195CPE7 A0A151XJ37 A0A194PRU4 A0A0F6Q2Z9 A0A291P0P8 A0A0F6Q4N3 A0A195FMJ1 A0A0M9A857 A0A1V0M879 A0A2A4JXJ6 A0A182VXI1 U5EPY7 E9ID65 A0A1S4G1B4 A0A182REU5 A0A182V6X2 A0A182I9C8 W5JHR7 A0A182JIX2 Q16GQ4 A0A195EP06 A0A182WVP9 A0A182LXW7 F4WVE2 Q7QAN3 A0A1B0CRT6 A0A0C9QUP1 A0A182KXM4 A0A182FRU7 A0A182QQN9 A0A182NFQ6 A0A1Q3G5B4 A0A182K6D3 A0A1W4XSR6 A0A1W4XHK8 A0A182P917 A0A1L8DYA8 A0A026WTB2 A0A182YI99 A0A084WSE9 A0A067RIF0 A0A1B6C0C4 E2AYW1 A0A195BV13 A0A182SVN7 A0A1B6J616 A0A212FLN3 A0A1Y1KTF8 A0A1B6FF15 A0A1A9UZB6 U4UHU2 A0A034W6Z8 N6TW75 A0A0P8ZQ48 W8BBB7 E0W0P6 B3MC49 B4KQR5 A0A2A3EMS1 A0A0R1DU59 A0A0K8WH42 A0A0K8UGQ7 A0A0Q5W393 B4J9N1 A0A336KQE3 D6WD88 B4PBX4 A0A0Q5VPH3 A0A0J9RKV0 A0A1B6DPR8 A0A0R3NKW5 A0A0J9RL31 A0A0A1XER0 Q8MMC6 Q0E8W1
U5KG45 A0A068FMK7 A0A1C9EGN8 A0A0F6Q2P8 A0A212EPZ8 I4DM82 A0A194QKX4 A0A194PEZ0 A0A088APG8 A0A3S2NWX2 D9MX50 A0A194QW79 A0A0L7QMT0 A0A0P0DZ32 A0A2H1VBF1 A0A0C9RGI8 A0A2W1BU73 A0A195CPE7 A0A151XJ37 A0A194PRU4 A0A0F6Q2Z9 A0A291P0P8 A0A0F6Q4N3 A0A195FMJ1 A0A0M9A857 A0A1V0M879 A0A2A4JXJ6 A0A182VXI1 U5EPY7 E9ID65 A0A1S4G1B4 A0A182REU5 A0A182V6X2 A0A182I9C8 W5JHR7 A0A182JIX2 Q16GQ4 A0A195EP06 A0A182WVP9 A0A182LXW7 F4WVE2 Q7QAN3 A0A1B0CRT6 A0A0C9QUP1 A0A182KXM4 A0A182FRU7 A0A182QQN9 A0A182NFQ6 A0A1Q3G5B4 A0A182K6D3 A0A1W4XSR6 A0A1W4XHK8 A0A182P917 A0A1L8DYA8 A0A026WTB2 A0A182YI99 A0A084WSE9 A0A067RIF0 A0A1B6C0C4 E2AYW1 A0A195BV13 A0A182SVN7 A0A1B6J616 A0A212FLN3 A0A1Y1KTF8 A0A1B6FF15 A0A1A9UZB6 U4UHU2 A0A034W6Z8 N6TW75 A0A0P8ZQ48 W8BBB7 E0W0P6 B3MC49 B4KQR5 A0A2A3EMS1 A0A0R1DU59 A0A0K8WH42 A0A0K8UGQ7 A0A0Q5W393 B4J9N1 A0A336KQE3 D6WD88 B4PBX4 A0A0Q5VPH3 A0A0J9RKV0 A0A1B6DPR8 A0A0R3NKW5 A0A0J9RL31 A0A0A1XER0 Q8MMC6 Q0E8W1
PDB
4W4T
E-value=1.61539e-08,
Score=143
Ontologies
PANTHER
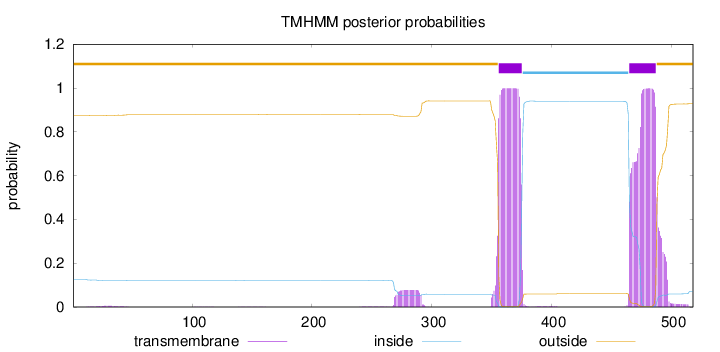
Topology
Length:
518
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.76507
Exp number, first 60 AAs:
0.11089
Total prob of N-in:
0.12497
outside
1 - 355
TMhelix
356 - 375
inside
376 - 464
TMhelix
465 - 487
outside
488 - 518
Population Genetic Test Statistics
Pi
227.077464
Theta
162.077106
Tajima's D
1.343033
CLR
0.003158
CSRT
0.750462476876156
Interpretation
Uncertain
