Gene
KWMTBOMO05601
Pre Gene Modal
BGIBMGA006572
Annotation
PREDICTED:_patched_domain-containing_protein_3_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.675
Sequence
CDS
ATGGGTTGTTGCAAACTGACAATCGTTGATGATGTCCTGAACAGGTCTTTCTACAAACTGGGCCTGGCCGTCGGCAGACAGCCCGGCTACTTCATAATAATACCGGTACTACTAACCTTGCTCATGGTAACCGGATATCAAAGGATTCATTATGAGATGGACCCGGAATATTTGTTTTCGCCTGTTAGCGGACAAGGAAAGTTGGAAAGGAGGGTAGTTGAAGAGCACTTCAAAGTTAACTACTCGCACAGATTTAACGTTGGCAGAATTACACGAGCAGGAAGATTTGGTCGAGTAATTATAGTGGCCAAAGACAACAATACTAACTTACTGCGAACCGAAATATGGAAAGAATTAAGGGAATTGGATGATCTGGTGCAAAACGTAACAGTCACGTTACCGACTGGAGAGACTTTCACGTATCGTGAAGAGTGCGCCAGATGGGAGGGGCAATGCTTCGTCAACGACATACTGAACTTAGACAAAATAATAGGCGAGGTGGAGCGCGGCGAGTTAAATCTGACTTTCCCAATAATGTTCAATCCCGTGACGTGGGAAGCACATGCATTTCCAGTATACTTTGGAGGAAGTCAAGTGGTTGATGATGTTATAGTGTCAGTGCCAGCCGTGCAATTGGTGTGGTTTGTTAGAACGGATACTAAAATACAGGAACAAAGGGGTGCGGCGTGGGAAGACGCGTTTTTGGACGCGGTGGGTATCGCCGAAGATACTGGCCGATTCAAGCACATATCGGTGGCTCGATTCGCTTCACGAACTCTTGACCATGAACTCGAAAAGAACACAAGAACCGTGATACCGTTCTTTAGTTCTACTTTCATTTTGATGGGTGTATTTTCAGTCGTCACTTGTTTGATGGGTGATTGGGTGCGTTCGAAGCCTTGGTTAGGATTACTGGGTAACATATCAGCCGTTATGGCCACGGCCGCAGCATTTGGCTGCGCGATATACTTAGGCATCTCTTTCATTGGAATAAATTTAGCGGCACCCTTCCTAATGATTGGTATCGGCATTGATGATACTTTTGTGATGTTGGCTGCGTGGAGAAGGGCTTCCCCAAAACTACCGGTTCCGGAACGCATGGCATCTATGTTATCAGAAGCCGCTGTTTCCATTACGATAACGTCGGTAACGGACATGTTGTCATTTTTCATTGGAATCTTTTCCCCGTTTCCATCTGTACAAATATTCTGCATGTACTCAGGTCTGGCGGTGTGTTTCACTTTTGTTTGGCATCTAACATTCTTTGCCGGTTGCGTCGCAGTTTCGGGATATCGTGAAAAACAAAACAGACACACAATTACTATGCTCAAAGTTTTGCCTGAATCGAGAGCACGTAAAGAAGAAAAATCGTGGTTATACAGGGCTTTCTGCAGCGGGGGCATTGATCCAGCCGATCCCGATAATCCAATTGACAATAAGGAACATTGTATCATGGCCTTCTTCCGTACAACCATGGCCGATTTGTTGAACCATAACGGAGTTAAAGCTTTCGTAATAGTTATATTCCTACTGTACTTGGCTGGAGCCGGTTACGGTGTTACCAATCTGAAGGAAGGTTTGGAAAGAAGAAAGCTGTCTAAAGTGGATTCATACTCTGTCGAGTTCTTCGATAGGGAGGATCTTTATTACAGAGAATTTCCATACAGGATACAGGTGGTAATTAGCGGTCAATACAATTATTCAGATACTTTAGTACAGGCTGAAGTTGAAAAACTGACAAAACGTTTGGAAAGCACATCGTACATATCGAATTCACTGTACACGGAGTCGTGGCTTAAAACGTTTTTGGATTACGTATCTGCTAATAACGACTATTTGAATGTGACTATCGATAACGAAGTTGACTTCATACAGAATCTCAAAGAGTTGTGGCTGTTTCCCGCTAATCCTTTCTCATTGGATGTCAAATTTAACGACGCCGGAGACCAAATCGTTGCTTCCAGGTTTCTGATACAAGCTATCAATATAAGTGGTACAAACCATGAAAAGGAAATGGTCAAAGCGTTAAGAACTATAGTTGCTGAATCACCGTTGAATGCTTCAGTGTTCCATCCGTATTTCGTATTTTTCGACCAGTTCGAGCTTGTTAAGCCTACATCAATTCAAAATTTGTGCTATGGAGCATTAATGATGATGATTACATCATTTATATTCATACCAAACATCTTGTGCTCGCTATGGGTGGCGTTTAGTATAATATCGATTGAGGTCGGTGTGGTCGGTTATATGGCCCTATGGGATATAAATCTCGATTCAATATCAATGATCAATCTCATCATGTGCATCGGTTTTTCTGTCGACTTCACTGCACATATATGCTATGCCTACATGGCATCTAAAGCGAAATACCCTCACGAACGAGTCAGCGAATGCTTATACTCTCTGGGATTACCAATAGTGCAAGGCTCTTTCAGCACAATTCTCGGTGTCATCGCATTGCTTCTCGCTGACAGTTACATATTCTCTGTATTCTTTAAAATGGTATTCATGGTAATATTCTTTGGAGCGATGCACGGTCTATTCTTATTACCGGTTTTACTGTCTTTGTTCGGACCAGGATCCTGCACAAGAAAATCAGACGATATGAAGATGTCGAAACTCGAAAAGTTTCCTAGCCCGTATTGTATACCGCACCCTCAGCTGATGTTGAACGATCAAATTTACAACAGCAAAAATTTGACCCCTAACGGAGTTTATAAAGGTTACGTGGACGATAAAGACCTGGGTATAGGGACGTCAGGCGAAGACACAAGTGAAAGCAGCTCTAATCAATCACAAAGACGACAAATCACCGAAAATGACAACGGACGCAAGCAATACGATGAAACAGGTTGGAAAAGATCGAATTATTGCCAAAGTTCGAGCCAATTTCAACCCTCCGGAGAATTAGATCCTTTCGGGCAAGACGTAGAAAGGAACTGGCATCGACAGCGCTATGAAAATAACCGTCGCACTTTTGAGAATTACGGTAAGCCTACACCCATTTCGAGGACCAGAGACGATGATTGGGTAACATCAACACGCAAAACCAGTGACACTGGACCTCGTCGAGAGGGTACATATAGAGTAATGCGGGCGCATTCACATCACAACCTCCACAGGCCCAGAGCTCCTCGTCGGTCTAACTCTCAGCATAATCTGGATCATTTAGATTACGTCGCAGAAATGCGATTTCCCTGA
Protein
MGCCKLTIVDDVLNRSFYKLGLAVGRQPGYFIIIPVLLTLLMVTGYQRIHYEMDPEYLFSPVSGQGKLERRVVEEHFKVNYSHRFNVGRITRAGRFGRVIIVAKDNNTNLLRTEIWKELRELDDLVQNVTVTLPTGETFTYREECARWEGQCFVNDILNLDKIIGEVERGELNLTFPIMFNPVTWEAHAFPVYFGGSQVVDDVIVSVPAVQLVWFVRTDTKIQEQRGAAWEDAFLDAVGIAEDTGRFKHISVARFASRTLDHELEKNTRTVIPFFSSTFILMGVFSVVTCLMGDWVRSKPWLGLLGNISAVMATAAAFGCAIYLGISFIGINLAAPFLMIGIGIDDTFVMLAAWRRASPKLPVPERMASMLSEAAVSITITSVTDMLSFFIGIFSPFPSVQIFCMYSGLAVCFTFVWHLTFFAGCVAVSGYREKQNRHTITMLKVLPESRARKEEKSWLYRAFCSGGIDPADPDNPIDNKEHCIMAFFRTTMADLLNHNGVKAFVIVIFLLYLAGAGYGVTNLKEGLERRKLSKVDSYSVEFFDREDLYYREFPYRIQVVISGQYNYSDTLVQAEVEKLTKRLESTSYISNSLYTESWLKTFLDYVSANNDYLNVTIDNEVDFIQNLKELWLFPANPFSLDVKFNDAGDQIVASRFLIQAINISGTNHEKEMVKALRTIVAESPLNASVFHPYFVFFDQFELVKPTSIQNLCYGALMMMITSFIFIPNILCSLWVAFSIISIEVGVVGYMALWDINLDSISMINLIMCIGFSVDFTAHICYAYMASKAKYPHERVSECLYSLGLPIVQGSFSTILGVIALLLADSYIFSVFFKMVFMVIFFGAMHGLFLLPVLLSLFGPGSCTRKSDDMKMSKLEKFPSPYCIPHPQLMLNDQIYNSKNLTPNGVYKGYVDDKDLGIGTSGEDTSESSSNQSQRRQITENDNGRKQYDETGWKRSNYCQSSSQFQPSGELDPFGQDVERNWHRQRYENNRRTFENYGKPTPISRTRDDDWVTSTRKTSDTGPRREGTYRVMRAHSHHNLHRPRAPRRSNSQHNLDHLDYVAEMRFP
Summary
Uniprot
A0A1L2D6R5
A0A2H1WTD7
A0A2A4JY65
A0A194PEZ9
A0A212FKD3
A0A1C9EGM5
+ More
A0A194QRM7 A0A0L7LDD9 H9JAM7 Q17LN2 A0A084W6U5 A0A182Q3L7 A0A023EWC3 A0A1S4H4P3 Q7PRH6 A0A139WCA8 A0A1Y1NIC1 A0A336K2U8 A0A0K8UBW4 A0A2J7QI84 A0A0L0BTK5 A0A1I8NTH2 W8B1J6 A0A1B0BX71 A0A1A9V1S5 A0A067QLA5 B4J489 B4MR35 B4NYU3 A0A1B0AG94 A0A0J9R699 Q86P36 B4LIL7 B4II44 B3NB69 A0A1W4V2B9 A0A0M4EWW5 B4KUI3 A0A0R1DVD4 B3MJ63 A0A1J1I1H5 B5E1M9 A0A158P0Z3 A0A195AUN0 B4HBD3 F4WTY0 A0A3B0JL01 A0A151J1H4 A0A151IPA3 A0A1B6CYR1 A0A0T6AWY4 A0A0L7QSL4 A0A151WWG6 A0A154PDC9 A0A151JUM0 A0A023F4K6 A0A0V0G2T1 A0A069DXM1 A4GVR7 A0A3L8E3A2 A0A224XE10 E2BA38 A0A0P4VN91 T1H8R3 J9JQ05 E1ZW52 A0A2A3EFB6 A0A0A9X8T6 A0A2H8TP81 A0A146L5X5 A0A1B6JHV1 A0A1S3DUH4 A0A0C9QXC8 N6SYM9 A0A2P8YWU5 K7IPV6 A0A232F287 A0A1B0G727 E0W0L7 E9IAD8 A0A1A9XIF1 A0A1A9WQZ3 B4QD75 A0A3B0J2I8 W8BB82 A0A087ZSR5 A0A2R7X378 A0A0P4VZS3 A0A1D2MK48 A0A226E2D1 A0A2P2HXT6 A0A3S4R506 A0A0N0BH07 A0A0K2T5I5 T1GSJ1
A0A194QRM7 A0A0L7LDD9 H9JAM7 Q17LN2 A0A084W6U5 A0A182Q3L7 A0A023EWC3 A0A1S4H4P3 Q7PRH6 A0A139WCA8 A0A1Y1NIC1 A0A336K2U8 A0A0K8UBW4 A0A2J7QI84 A0A0L0BTK5 A0A1I8NTH2 W8B1J6 A0A1B0BX71 A0A1A9V1S5 A0A067QLA5 B4J489 B4MR35 B4NYU3 A0A1B0AG94 A0A0J9R699 Q86P36 B4LIL7 B4II44 B3NB69 A0A1W4V2B9 A0A0M4EWW5 B4KUI3 A0A0R1DVD4 B3MJ63 A0A1J1I1H5 B5E1M9 A0A158P0Z3 A0A195AUN0 B4HBD3 F4WTY0 A0A3B0JL01 A0A151J1H4 A0A151IPA3 A0A1B6CYR1 A0A0T6AWY4 A0A0L7QSL4 A0A151WWG6 A0A154PDC9 A0A151JUM0 A0A023F4K6 A0A0V0G2T1 A0A069DXM1 A4GVR7 A0A3L8E3A2 A0A224XE10 E2BA38 A0A0P4VN91 T1H8R3 J9JQ05 E1ZW52 A0A2A3EFB6 A0A0A9X8T6 A0A2H8TP81 A0A146L5X5 A0A1B6JHV1 A0A1S3DUH4 A0A0C9QXC8 N6SYM9 A0A2P8YWU5 K7IPV6 A0A232F287 A0A1B0G727 E0W0L7 E9IAD8 A0A1A9XIF1 A0A1A9WQZ3 B4QD75 A0A3B0J2I8 W8BB82 A0A087ZSR5 A0A2R7X378 A0A0P4VZS3 A0A1D2MK48 A0A226E2D1 A0A2P2HXT6 A0A3S4R506 A0A0N0BH07 A0A0K2T5I5 T1GSJ1
Pubmed
26354079
22118469
26227816
19121390
17510324
24438588
+ More
24945155 12364791 18362917 19820115 28004739 26108605 24495485 24845553 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 21347285 21719571 25474469 26334808 18563713 30249741 20798317 27129103 25401762 26823975 23537049 29403074 20075255 28648823 20566863 21282665 27289101
24945155 12364791 18362917 19820115 28004739 26108605 24495485 24845553 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 21347285 21719571 25474469 26334808 18563713 30249741 20798317 27129103 25401762 26823975 23537049 29403074 20075255 28648823 20566863 21282665 27289101
EMBL
KR338939
AMR74905.1
ODYU01010914
SOQ56318.1
NWSH01000389
PCG76749.1
+ More
KQ459606 KPI91598.1 AGBW02008063 OWR54203.1 KX778610 AON96648.1 KQ461198 KPJ06191.1 JTDY01001599 KOB73414.1 BABH01008690 BABH01008691 CH477214 EAT47579.1 ATLV01020950 ATLV01020951 ATLV01020952 ATLV01020953 KE525310 KFB45939.1 AXCN02002295 GAPW01000068 JAC13530.1 AAAB01008849 EAA07199.5 KQ971371 KYB25536.1 GEZM01001584 JAV97652.1 UFQS01000061 UFQT01000061 SSW98787.1 SSX19173.1 GDHF01028499 JAI23815.1 NEVH01013957 PNF28292.1 JRES01001352 KNC23395.1 GAMC01019450 GAMC01019449 JAB87106.1 JXJN01022098 KK853300 KDR08758.1 CH916367 EDW00569.1 CH963849 EDW74574.1 CM000157 EDW89794.1 CM002911 KMY91712.1 AE013599 BT003500 AAF57274.2 AAO39504.1 AFH07900.1 AFH07901.1 CH940648 EDW60387.2 CH480841 EDW49570.1 CH954177 EDV59834.1 CP012524 ALC42675.1 CH933808 EDW10770.2 KRG05467.1 KRJ98710.1 CH902619 EDV38157.1 CVRI01000038 CRK94133.1 CM000071 EDY69532.2 ADTU01005915 ADTU01005916 ADTU01005917 KQ976738 KYM75767.1 CH479252 EDW38675.1 GL888345 EGI62337.1 OUUW01000001 SPP73321.1 KQ980514 KYN15729.1 KQ976878 KYN07667.1 GEDC01018770 JAS18528.1 LJIG01022611 KRT79642.1 KQ414756 KOC61632.1 KQ982691 KYQ52156.1 KQ434868 KZC09210.1 KQ981732 KYN36560.1 GBBI01002500 JAC16212.1 GECL01003692 JAP02432.1 GBGD01000318 JAC88571.1 EF442429 ABO28476.1 QOIP01000001 RLU27167.1 GFTR01008398 JAW08028.1 GL446664 EFN87447.1 GDKW01002634 JAI53961.1 ACPB03020617 ABLF02028363 ABLF02028367 GL434776 EFN74561.1 KZ288266 PBC30174.1 GBHO01030090 GBHO01030089 JAG13514.1 JAG13515.1 GFXV01004014 MBW15819.1 GDHC01016003 JAQ02626.1 GECU01008922 JAS98784.1 GBYB01005302 JAG75069.1 APGK01058856 APGK01058857 APGK01058858 APGK01058859 APGK01058860 KB741292 ENN70323.1 PYGN01000312 PSN48726.1 NNAY01001161 OXU24936.1 CCAG010003764 DS235861 EEB19173.1 GL762003 EFZ22481.1 CM000362 EDX05862.1 SPP73322.1 GAMC01019451 JAB87104.1 KK856273 PTY25355.1 GDRN01095418 JAI59530.1 LJIJ01001034 ODM93265.1 LNIX01000007 OXA51440.1 IACF01000676 LAB66436.1 NCKU01001607 NCKU01001606 NCKU01001540 NCKU01001539 RWS11672.1 RWS11674.1 RWS11862.1 RWS11869.1 KQ435760 KOX75554.1 HACA01003355 CDW20716.1 CAQQ02129062 CAQQ02129063 CAQQ02129064
KQ459606 KPI91598.1 AGBW02008063 OWR54203.1 KX778610 AON96648.1 KQ461198 KPJ06191.1 JTDY01001599 KOB73414.1 BABH01008690 BABH01008691 CH477214 EAT47579.1 ATLV01020950 ATLV01020951 ATLV01020952 ATLV01020953 KE525310 KFB45939.1 AXCN02002295 GAPW01000068 JAC13530.1 AAAB01008849 EAA07199.5 KQ971371 KYB25536.1 GEZM01001584 JAV97652.1 UFQS01000061 UFQT01000061 SSW98787.1 SSX19173.1 GDHF01028499 JAI23815.1 NEVH01013957 PNF28292.1 JRES01001352 KNC23395.1 GAMC01019450 GAMC01019449 JAB87106.1 JXJN01022098 KK853300 KDR08758.1 CH916367 EDW00569.1 CH963849 EDW74574.1 CM000157 EDW89794.1 CM002911 KMY91712.1 AE013599 BT003500 AAF57274.2 AAO39504.1 AFH07900.1 AFH07901.1 CH940648 EDW60387.2 CH480841 EDW49570.1 CH954177 EDV59834.1 CP012524 ALC42675.1 CH933808 EDW10770.2 KRG05467.1 KRJ98710.1 CH902619 EDV38157.1 CVRI01000038 CRK94133.1 CM000071 EDY69532.2 ADTU01005915 ADTU01005916 ADTU01005917 KQ976738 KYM75767.1 CH479252 EDW38675.1 GL888345 EGI62337.1 OUUW01000001 SPP73321.1 KQ980514 KYN15729.1 KQ976878 KYN07667.1 GEDC01018770 JAS18528.1 LJIG01022611 KRT79642.1 KQ414756 KOC61632.1 KQ982691 KYQ52156.1 KQ434868 KZC09210.1 KQ981732 KYN36560.1 GBBI01002500 JAC16212.1 GECL01003692 JAP02432.1 GBGD01000318 JAC88571.1 EF442429 ABO28476.1 QOIP01000001 RLU27167.1 GFTR01008398 JAW08028.1 GL446664 EFN87447.1 GDKW01002634 JAI53961.1 ACPB03020617 ABLF02028363 ABLF02028367 GL434776 EFN74561.1 KZ288266 PBC30174.1 GBHO01030090 GBHO01030089 JAG13514.1 JAG13515.1 GFXV01004014 MBW15819.1 GDHC01016003 JAQ02626.1 GECU01008922 JAS98784.1 GBYB01005302 JAG75069.1 APGK01058856 APGK01058857 APGK01058858 APGK01058859 APGK01058860 KB741292 ENN70323.1 PYGN01000312 PSN48726.1 NNAY01001161 OXU24936.1 CCAG010003764 DS235861 EEB19173.1 GL762003 EFZ22481.1 CM000362 EDX05862.1 SPP73322.1 GAMC01019451 JAB87104.1 KK856273 PTY25355.1 GDRN01095418 JAI59530.1 LJIJ01001034 ODM93265.1 LNIX01000007 OXA51440.1 IACF01000676 LAB66436.1 NCKU01001607 NCKU01001606 NCKU01001540 NCKU01001539 RWS11672.1 RWS11674.1 RWS11862.1 RWS11869.1 KQ435760 KOX75554.1 HACA01003355 CDW20716.1 CAQQ02129062 CAQQ02129063 CAQQ02129064
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
UP000005204
+ More
UP000008820 UP000030765 UP000075886 UP000007062 UP000007266 UP000235965 UP000037069 UP000095300 UP000092460 UP000078200 UP000027135 UP000001070 UP000007798 UP000002282 UP000092445 UP000000803 UP000008792 UP000001292 UP000008711 UP000192221 UP000092553 UP000009192 UP000007801 UP000183832 UP000001819 UP000005205 UP000078540 UP000008744 UP000007755 UP000268350 UP000078492 UP000078542 UP000053825 UP000075809 UP000076502 UP000078541 UP000279307 UP000008237 UP000015103 UP000007819 UP000000311 UP000242457 UP000079169 UP000019118 UP000245037 UP000002358 UP000215335 UP000092444 UP000009046 UP000092443 UP000091820 UP000000304 UP000005203 UP000094527 UP000198287 UP000285301 UP000053105 UP000015102
UP000008820 UP000030765 UP000075886 UP000007062 UP000007266 UP000235965 UP000037069 UP000095300 UP000092460 UP000078200 UP000027135 UP000001070 UP000007798 UP000002282 UP000092445 UP000000803 UP000008792 UP000001292 UP000008711 UP000192221 UP000092553 UP000009192 UP000007801 UP000183832 UP000001819 UP000005205 UP000078540 UP000008744 UP000007755 UP000268350 UP000078492 UP000078542 UP000053825 UP000075809 UP000076502 UP000078541 UP000279307 UP000008237 UP000015103 UP000007819 UP000000311 UP000242457 UP000079169 UP000019118 UP000245037 UP000002358 UP000215335 UP000092444 UP000009046 UP000092443 UP000091820 UP000000304 UP000005203 UP000094527 UP000198287 UP000285301 UP000053105 UP000015102
ProteinModelPortal
A0A1L2D6R5
A0A2H1WTD7
A0A2A4JY65
A0A194PEZ9
A0A212FKD3
A0A1C9EGM5
+ More
A0A194QRM7 A0A0L7LDD9 H9JAM7 Q17LN2 A0A084W6U5 A0A182Q3L7 A0A023EWC3 A0A1S4H4P3 Q7PRH6 A0A139WCA8 A0A1Y1NIC1 A0A336K2U8 A0A0K8UBW4 A0A2J7QI84 A0A0L0BTK5 A0A1I8NTH2 W8B1J6 A0A1B0BX71 A0A1A9V1S5 A0A067QLA5 B4J489 B4MR35 B4NYU3 A0A1B0AG94 A0A0J9R699 Q86P36 B4LIL7 B4II44 B3NB69 A0A1W4V2B9 A0A0M4EWW5 B4KUI3 A0A0R1DVD4 B3MJ63 A0A1J1I1H5 B5E1M9 A0A158P0Z3 A0A195AUN0 B4HBD3 F4WTY0 A0A3B0JL01 A0A151J1H4 A0A151IPA3 A0A1B6CYR1 A0A0T6AWY4 A0A0L7QSL4 A0A151WWG6 A0A154PDC9 A0A151JUM0 A0A023F4K6 A0A0V0G2T1 A0A069DXM1 A4GVR7 A0A3L8E3A2 A0A224XE10 E2BA38 A0A0P4VN91 T1H8R3 J9JQ05 E1ZW52 A0A2A3EFB6 A0A0A9X8T6 A0A2H8TP81 A0A146L5X5 A0A1B6JHV1 A0A1S3DUH4 A0A0C9QXC8 N6SYM9 A0A2P8YWU5 K7IPV6 A0A232F287 A0A1B0G727 E0W0L7 E9IAD8 A0A1A9XIF1 A0A1A9WQZ3 B4QD75 A0A3B0J2I8 W8BB82 A0A087ZSR5 A0A2R7X378 A0A0P4VZS3 A0A1D2MK48 A0A226E2D1 A0A2P2HXT6 A0A3S4R506 A0A0N0BH07 A0A0K2T5I5 T1GSJ1
A0A194QRM7 A0A0L7LDD9 H9JAM7 Q17LN2 A0A084W6U5 A0A182Q3L7 A0A023EWC3 A0A1S4H4P3 Q7PRH6 A0A139WCA8 A0A1Y1NIC1 A0A336K2U8 A0A0K8UBW4 A0A2J7QI84 A0A0L0BTK5 A0A1I8NTH2 W8B1J6 A0A1B0BX71 A0A1A9V1S5 A0A067QLA5 B4J489 B4MR35 B4NYU3 A0A1B0AG94 A0A0J9R699 Q86P36 B4LIL7 B4II44 B3NB69 A0A1W4V2B9 A0A0M4EWW5 B4KUI3 A0A0R1DVD4 B3MJ63 A0A1J1I1H5 B5E1M9 A0A158P0Z3 A0A195AUN0 B4HBD3 F4WTY0 A0A3B0JL01 A0A151J1H4 A0A151IPA3 A0A1B6CYR1 A0A0T6AWY4 A0A0L7QSL4 A0A151WWG6 A0A154PDC9 A0A151JUM0 A0A023F4K6 A0A0V0G2T1 A0A069DXM1 A4GVR7 A0A3L8E3A2 A0A224XE10 E2BA38 A0A0P4VN91 T1H8R3 J9JQ05 E1ZW52 A0A2A3EFB6 A0A0A9X8T6 A0A2H8TP81 A0A146L5X5 A0A1B6JHV1 A0A1S3DUH4 A0A0C9QXC8 N6SYM9 A0A2P8YWU5 K7IPV6 A0A232F287 A0A1B0G727 E0W0L7 E9IAD8 A0A1A9XIF1 A0A1A9WQZ3 B4QD75 A0A3B0J2I8 W8BB82 A0A087ZSR5 A0A2R7X378 A0A0P4VZS3 A0A1D2MK48 A0A226E2D1 A0A2P2HXT6 A0A3S4R506 A0A0N0BH07 A0A0K2T5I5 T1GSJ1
PDB
5JNX
E-value=8.0696e-29,
Score=321
Ontologies
GO
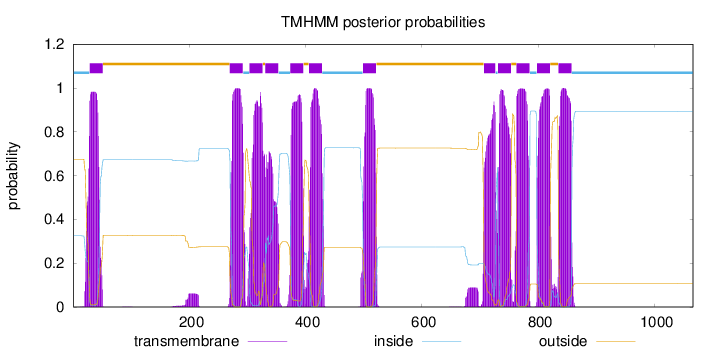
Topology
Length:
1066
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
255.53712
Exp number, first 60 AAs:
21.00837
Total prob of N-in:
0.32538
POSSIBLE N-term signal
sequence
inside
1 - 28
TMhelix
29 - 51
outside
52 - 269
TMhelix
270 - 292
inside
293 - 303
TMhelix
304 - 326
outside
327 - 330
TMhelix
331 - 353
inside
354 - 373
TMhelix
374 - 396
outside
397 - 405
TMhelix
406 - 428
inside
429 - 498
TMhelix
499 - 521
outside
522 - 706
TMhelix
707 - 726
inside
727 - 730
TMhelix
731 - 753
outside
754 - 762
TMhelix
763 - 785
inside
786 - 797
TMhelix
798 - 820
outside
821 - 834
TMhelix
835 - 857
inside
858 - 1066
Population Genetic Test Statistics
Pi
203.598931
Theta
181.680595
Tajima's D
0.360462
CLR
0.500524
CSRT
0.469176541172941
Interpretation
Uncertain
