Pre Gene Modal
BGIBMGA006574
Annotation
heat_shock_70_kDa_protein_14-like_[Bombyx_mori]
Full name
Heat shock 70 kDa protein 14
Location in the cell
Cytoplasmic Reliability : 1.707 Golgi Reliability : 1.238
Sequence
CDS
ATGGCATCAGCTTACGGAATACATTTGGGTAACAGTACAGCATGCTTAGCATCGTTTACGAACGGCACAATATCTGTGATAGCCAATGACGCCGGCGATCGTGTCACCCCCGCAATAATTGCTCTAAATGGTACAGAATGGGAAATTGGCCTTCCAGCAAAGTCAGGACAAGCGTCATCGAAGTCAATTATTAAGCACAATAAACGGCTAATGAACTGTGATTTCAATGAAGATGATTTGTCATATGTTGAGTTATCATCATCGTGCCAAATTCAGAACGACGAAAAACTTGTATATGAATTCCAGACAAGCGAAACAACATTATACTCTAGTCCTGATAACATCGCCACTAAAATTTATGCTAAACTATTTACGATTGCTAGTCATTCTGTGAAGAATGAGGGTGATCTCAAATTGGTGCTCGCTGCCCCACTACATTGGTCAAATGAAAGCAGAGAACGCTTAGTAAAATGCGCAGAACTGGCGGGATTCGAAGTACTTCAAGTGATTAGTGAACCGGCTGCTGCTTTATTAGCCTATAACATTGAAGAAAATCTAGAAGATGTGAATGTACTTATTTATCGACTTGGTGGATCATCTTGTGATGTATCTATTGCCAAAGTATCAGCTGGTTTTGTTAGTGTTGAAAAAAATATTTTTCGTTCGGATTTAGGCGGACAGTACTTGACAAAAGATCTTGCTGATTTTATTGCACAAGAATTTAAACAGAAGTGGAAGTTAGACCCTCAGGAAAGTAAAAGAGCCATGGCCAAACTTTTAAACCATGCTGATAACTGTAAAAACGTTTTATCTACTTTAAATTCAGCTCATGTATTTATTGAGTCATTGCTTGATGGTGTTGACTGGAGCCAAAATGTGTCTCGGGCTAGATTTGAGAACATAATATCATCAAAAATAGCTTCCTATGTAGAGCCTGCCGAGAAGCTAATACAAAGTTTTGATGGTAAAATTAGCAAAATTGTGTTATGTGGAGGGAGTATGAAGATACCTAAATTACAGTCTACCATTGCAAATTTGATTCCTGATGCTGAAGTTCTGTCGGGTATAAACCCTGATGAGGTAATAGCTATTGGTTGTGCTAAAGAGGCAGGGTTGTTATTAGATGTTCCAGAATTTTCTATGAACGATTTAAATACAGAAGTTGAATTCTTAGGCAAAGATGTGTATATGAAATATTTAGATCAAACAGTCCACCTGTTTAAGGAAGGTACACCTCCATTTGCACAATTTGTTCAAAGTGTGGAAGCAGAGCAAGAAATAAAAGATATTGATTTCACCTTACACACATCACCTGAAAGTGAGGCTTATGCTACAGAATCATTCAATATTGAGAATCTTAAAAAACCTTTTTCATTAAAAGCAACATTACAACAATCAAGTATAAAATTGGAAGTTGAATAA
Protein
MASAYGIHLGNSTACLASFTNGTISVIANDAGDRVTPAIIALNGTEWEIGLPAKSGQASSKSIIKHNKRLMNCDFNEDDLSYVELSSSCQIQNDEKLVYEFQTSETTLYSSPDNIATKIYAKLFTIASHSVKNEGDLKLVLAAPLHWSNESRERLVKCAELAGFEVLQVISEPAAALLAYNIEENLEDVNVLIYRLGGSSCDVSIAKVSAGFVSVEKNIFRSDLGGQYLTKDLADFIAQEFKQKWKLDPQESKRAMAKLLNHADNCKNVLSTLNSAHVFIESLLDGVDWSQNVSRARFENIISSKIASYVEPAEKLIQSFDGKISKIVLCGGSMKIPKLQSTIANLIPDAEVLSGINPDEVIAIGCAKEAGLLLDVPEFSMNDLNTEVEFLGKDVYMKYLDQTVHLFKEGTPPFAQFVQSVEAEQEIKDIDFTLHTSPESEAYATESFNIENLKKPFSLKATLQQSSIKLEVE
Summary
Description
Component of the ribosome-associated complex (RAC), a complex involved in folding or maintaining nascent polypeptides in a folding-competent state.
Component of the ribosome-associated complex (RAC), a complex involved in folding or maintaining nascent polypeptides in a folding-competent state. In the RAC complex, binds to the nascent polypeptide chain, while DNAJC2 stimulates its ATPase activity (By similarity).
Component of the ribosome-associated complex (RAC), a complex involved in folding or maintaining nascent polypeptides in a folding-competent state. In the RAC complex, binds to the nascent polypeptide chain, while DNAJC2 stimulates its ATPase activity (By similarity).
Subunit
Component of ribosome-associated complex (RAC).
Component of ribosome-associated complex (RAC), a heterodimer composed of Hsp70/DnaK-type chaperone HSPA14 and Hsp40/DnaJ-type chaperone DNAJC2.
Component of ribosome-associated complex (RAC), a heterodimer composed of Hsp70/DnaK-type chaperone HSPA14 and Hsp40/DnaJ-type chaperone DNAJC2.
Similarity
Belongs to the heat shock protein 70 family.
Keywords
ATP-binding
Chaperone
Complete proteome
Cytoplasm
Nucleotide-binding
Reference proteome
Feature
chain Heat shock 70 kDa protein 14
Uniprot
H9JAM9
A0A2H1WS87
A0A2A4JYM6
A0A0L7LFY5
A0A1C9EGM3
A0A212FKD2
+ More
A0A194QML0 I4DPQ0 A0A194PF04 A0A2J7QBD4 A0A067RF33 A0A2P8Y1Z2 U5ESP3 A0A0N7ZSD9 A0A0P4Z6I9 A0A0P4ZQS6 A0A0P5R4A2 A0A164RPP1 E9GX91 A0A0C9RY66 A0A1S3JS87 Q5RGE6 A0A2R8QL69 A0A3Q1M9A5 H3AQ29 A0A087X5B2 W5NJ54 A0A3Q3BF84 A0A336MHC5 Q2YDD0 W5U834 A0A3P9PWY1 A0A3P9PWQ1 M3ZS18 L8INA2 A0A315V9H2 A0A3B3U5B4 A0A3B3VHG6 A0A1A8PB39 A0A1A8LVI3 A0A2I4BQK9 A0A2Y9KCJ1 A0A3Q7TFY1 A0A3Q7T552 A0A1A8A8L2 A0A3Q1H6G9 A0A2U3V2E4 E2RHI0 A0A2U3WS28 A0A3Q1C0N7 A0A3P8TDC0 A0A1A8V9N8 A0A1A8BE07 A0A3B5PXX6 A0A3Q2LPT5 A0A340XLE4 F7AUE0 A0A341AUR3 W5KQY9 A0A2Y9NYU7 A0A3Q0RC56 A0A2Y9NTG0 A0A1S3AAR9 A0A2Y9FLC5 A0A384B9Z4 A0A1A8JIH6 A0A1A8FRG3 A0A1A8J2G4 A0A2Y9HL33 M3WAW7 A0A1A8FGW6 A0A1A7YF47 A0A146MM25 I3K9M5 A0A3B4DNV6 A0A3Q7MQ43 G3SY46 A0A3Q2Z7K7 A0A2U3XV83 F7IA97 F1RTY7 A0A2K5D5F4 U3CHH0 U3BDK8 G1LYF4 A0A3B4ZE85 F6VN33 A0A3B4ZE58 A0A3Q3JVW0 G2HEZ2 A0A1S3F0L6 A0A3B3QW90 H2N9T6 A0A2U9BAY7 A0A2R8ZHF6 L5LUB0 A0A2J8VKL9 A0A3P8PW12
A0A194QML0 I4DPQ0 A0A194PF04 A0A2J7QBD4 A0A067RF33 A0A2P8Y1Z2 U5ESP3 A0A0N7ZSD9 A0A0P4Z6I9 A0A0P4ZQS6 A0A0P5R4A2 A0A164RPP1 E9GX91 A0A0C9RY66 A0A1S3JS87 Q5RGE6 A0A2R8QL69 A0A3Q1M9A5 H3AQ29 A0A087X5B2 W5NJ54 A0A3Q3BF84 A0A336MHC5 Q2YDD0 W5U834 A0A3P9PWY1 A0A3P9PWQ1 M3ZS18 L8INA2 A0A315V9H2 A0A3B3U5B4 A0A3B3VHG6 A0A1A8PB39 A0A1A8LVI3 A0A2I4BQK9 A0A2Y9KCJ1 A0A3Q7TFY1 A0A3Q7T552 A0A1A8A8L2 A0A3Q1H6G9 A0A2U3V2E4 E2RHI0 A0A2U3WS28 A0A3Q1C0N7 A0A3P8TDC0 A0A1A8V9N8 A0A1A8BE07 A0A3B5PXX6 A0A3Q2LPT5 A0A340XLE4 F7AUE0 A0A341AUR3 W5KQY9 A0A2Y9NYU7 A0A3Q0RC56 A0A2Y9NTG0 A0A1S3AAR9 A0A2Y9FLC5 A0A384B9Z4 A0A1A8JIH6 A0A1A8FRG3 A0A1A8J2G4 A0A2Y9HL33 M3WAW7 A0A1A8FGW6 A0A1A7YF47 A0A146MM25 I3K9M5 A0A3B4DNV6 A0A3Q7MQ43 G3SY46 A0A3Q2Z7K7 A0A2U3XV83 F7IA97 F1RTY7 A0A2K5D5F4 U3CHH0 U3BDK8 G1LYF4 A0A3B4ZE85 F6VN33 A0A3B4ZE58 A0A3Q3JVW0 G2HEZ2 A0A1S3F0L6 A0A3B3QW90 H2N9T6 A0A2U9BAY7 A0A2R8ZHF6 L5LUB0 A0A2J8VKL9 A0A3P8PW12
Pubmed
EMBL
BABH01008688
JX014251
AFN02501.1
ODYU01010635
SOQ55877.1
NWSH01000389
+ More
PCG76744.1 JTDY01001283 KOB74300.1 KX778610 AON96644.1 AGBW02008063 OWR54207.1 KQ461198 KPJ06195.1 AK403713 BAM19890.1 KQ459606 KPI91603.1 NEVH01016301 PNF25879.1 KK852729 KDR17583.1 PYGN01001036 PSN38263.1 GANO01003174 JAB56697.1 GDIP01217225 JAJ06177.1 GDIP01217224 JAJ06178.1 GDIP01222588 JAJ00814.1 GDIQ01115236 JAL36490.1 LRGB01002167 KZS08838.1 GL732572 EFX75829.1 GBYB01003685 GBYB01012871 JAG73452.1 JAG82638.1 BX901962 BC129343 BX470236 AFYH01187965 AFYH01187966 AFYH01187967 AFYH01187968 AYCK01021689 AHAT01023664 UFQT01001180 SSX29360.1 BC110281 JT406759 AHH37714.1 JH881041 ELR57024.1 NHOQ01002007 PWA20019.1 HAEI01002011 HAEH01016335 SBR78595.1 HAEF01009161 HAEG01009316 SBR47969.1 HADY01012972 SBP51457.1 AAEX03001258 HADY01018893 HAEJ01015840 SBS56297.1 HADZ01001523 HAEA01015370 SBP65464.1 HAED01022811 SBR09584.1 HAEB01015090 SBQ61617.1 HAED01017430 HAEE01011313 SBR03875.1 AANG04002256 HAEB01012133 HAEC01002639 SBQ58660.1 HADW01001637 HADX01006930 SBP29162.1 GCES01166210 JAQ20112.1 AERX01038201 AEMK02000074 GAMR01008864 JAB25068.1 GAMT01008137 GAMS01005338 GAMQ01004408 GAMP01005574 JAB03724.1 JAB17798.1 JAB37443.1 JAB47181.1 ACTA01040312 AACZ04052237 AACZ04052238 AACZ04052239 AK305306 GABC01007821 GABF01003948 GABD01009525 GABE01010145 NBAG03000034 BAK62300.1 JAA03517.1 JAA18197.1 JAA23575.1 JAA34594.1 PNI96425.1 ABGA01122710 ABGA01122711 ABGA01122712 ABGA01122713 ABGA01122714 ABGA01122715 ABGA01122716 CP026246 AWP00986.1 AJFE02019012 AJFE02019013 AJFE02019014 AJFE02019015 KB108520 ELK29043.1 NDHI03003418 PNJ58057.1
PCG76744.1 JTDY01001283 KOB74300.1 KX778610 AON96644.1 AGBW02008063 OWR54207.1 KQ461198 KPJ06195.1 AK403713 BAM19890.1 KQ459606 KPI91603.1 NEVH01016301 PNF25879.1 KK852729 KDR17583.1 PYGN01001036 PSN38263.1 GANO01003174 JAB56697.1 GDIP01217225 JAJ06177.1 GDIP01217224 JAJ06178.1 GDIP01222588 JAJ00814.1 GDIQ01115236 JAL36490.1 LRGB01002167 KZS08838.1 GL732572 EFX75829.1 GBYB01003685 GBYB01012871 JAG73452.1 JAG82638.1 BX901962 BC129343 BX470236 AFYH01187965 AFYH01187966 AFYH01187967 AFYH01187968 AYCK01021689 AHAT01023664 UFQT01001180 SSX29360.1 BC110281 JT406759 AHH37714.1 JH881041 ELR57024.1 NHOQ01002007 PWA20019.1 HAEI01002011 HAEH01016335 SBR78595.1 HAEF01009161 HAEG01009316 SBR47969.1 HADY01012972 SBP51457.1 AAEX03001258 HADY01018893 HAEJ01015840 SBS56297.1 HADZ01001523 HAEA01015370 SBP65464.1 HAED01022811 SBR09584.1 HAEB01015090 SBQ61617.1 HAED01017430 HAEE01011313 SBR03875.1 AANG04002256 HAEB01012133 HAEC01002639 SBQ58660.1 HADW01001637 HADX01006930 SBP29162.1 GCES01166210 JAQ20112.1 AERX01038201 AEMK02000074 GAMR01008864 JAB25068.1 GAMT01008137 GAMS01005338 GAMQ01004408 GAMP01005574 JAB03724.1 JAB17798.1 JAB37443.1 JAB47181.1 ACTA01040312 AACZ04052237 AACZ04052238 AACZ04052239 AK305306 GABC01007821 GABF01003948 GABD01009525 GABE01010145 NBAG03000034 BAK62300.1 JAA03517.1 JAA18197.1 JAA23575.1 JAA34594.1 PNI96425.1 ABGA01122710 ABGA01122711 ABGA01122712 ABGA01122713 ABGA01122714 ABGA01122715 ABGA01122716 CP026246 AWP00986.1 AJFE02019012 AJFE02019013 AJFE02019014 AJFE02019015 KB108520 ELK29043.1 NDHI03003418 PNJ58057.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000235965 UP000027135 UP000245037 UP000076858 UP000000305 UP000085678 UP000000437 UP000009136 UP000008672 UP000028760 UP000018468 UP000264800 UP000221080 UP000242638 UP000002852 UP000261500 UP000192220 UP000248482 UP000286642 UP000286640 UP000265040 UP000245320 UP000002254 UP000245340 UP000257160 UP000265080 UP000002281 UP000265300 UP000252040 UP000018467 UP000248483 UP000261340 UP000079721 UP000248484 UP000261681 UP000248481 UP000011712 UP000005207 UP000261440 UP000286641 UP000007646 UP000264820 UP000245341 UP000008225 UP000008227 UP000233020 UP000008912 UP000261400 UP000002280 UP000261600 UP000002277 UP000081671 UP000261540 UP000001595 UP000246464 UP000240080 UP000265100
UP000235965 UP000027135 UP000245037 UP000076858 UP000000305 UP000085678 UP000000437 UP000009136 UP000008672 UP000028760 UP000018468 UP000264800 UP000221080 UP000242638 UP000002852 UP000261500 UP000192220 UP000248482 UP000286642 UP000286640 UP000265040 UP000245320 UP000002254 UP000245340 UP000257160 UP000265080 UP000002281 UP000265300 UP000252040 UP000018467 UP000248483 UP000261340 UP000079721 UP000248484 UP000261681 UP000248481 UP000011712 UP000005207 UP000261440 UP000286641 UP000007646 UP000264820 UP000245341 UP000008225 UP000008227 UP000233020 UP000008912 UP000261400 UP000002280 UP000261600 UP000002277 UP000081671 UP000261540 UP000001595 UP000246464 UP000240080 UP000265100
Interpro
Gene 3D
ProteinModelPortal
H9JAM9
A0A2H1WS87
A0A2A4JYM6
A0A0L7LFY5
A0A1C9EGM3
A0A212FKD2
+ More
A0A194QML0 I4DPQ0 A0A194PF04 A0A2J7QBD4 A0A067RF33 A0A2P8Y1Z2 U5ESP3 A0A0N7ZSD9 A0A0P4Z6I9 A0A0P4ZQS6 A0A0P5R4A2 A0A164RPP1 E9GX91 A0A0C9RY66 A0A1S3JS87 Q5RGE6 A0A2R8QL69 A0A3Q1M9A5 H3AQ29 A0A087X5B2 W5NJ54 A0A3Q3BF84 A0A336MHC5 Q2YDD0 W5U834 A0A3P9PWY1 A0A3P9PWQ1 M3ZS18 L8INA2 A0A315V9H2 A0A3B3U5B4 A0A3B3VHG6 A0A1A8PB39 A0A1A8LVI3 A0A2I4BQK9 A0A2Y9KCJ1 A0A3Q7TFY1 A0A3Q7T552 A0A1A8A8L2 A0A3Q1H6G9 A0A2U3V2E4 E2RHI0 A0A2U3WS28 A0A3Q1C0N7 A0A3P8TDC0 A0A1A8V9N8 A0A1A8BE07 A0A3B5PXX6 A0A3Q2LPT5 A0A340XLE4 F7AUE0 A0A341AUR3 W5KQY9 A0A2Y9NYU7 A0A3Q0RC56 A0A2Y9NTG0 A0A1S3AAR9 A0A2Y9FLC5 A0A384B9Z4 A0A1A8JIH6 A0A1A8FRG3 A0A1A8J2G4 A0A2Y9HL33 M3WAW7 A0A1A8FGW6 A0A1A7YF47 A0A146MM25 I3K9M5 A0A3B4DNV6 A0A3Q7MQ43 G3SY46 A0A3Q2Z7K7 A0A2U3XV83 F7IA97 F1RTY7 A0A2K5D5F4 U3CHH0 U3BDK8 G1LYF4 A0A3B4ZE85 F6VN33 A0A3B4ZE58 A0A3Q3JVW0 G2HEZ2 A0A1S3F0L6 A0A3B3QW90 H2N9T6 A0A2U9BAY7 A0A2R8ZHF6 L5LUB0 A0A2J8VKL9 A0A3P8PW12
A0A194QML0 I4DPQ0 A0A194PF04 A0A2J7QBD4 A0A067RF33 A0A2P8Y1Z2 U5ESP3 A0A0N7ZSD9 A0A0P4Z6I9 A0A0P4ZQS6 A0A0P5R4A2 A0A164RPP1 E9GX91 A0A0C9RY66 A0A1S3JS87 Q5RGE6 A0A2R8QL69 A0A3Q1M9A5 H3AQ29 A0A087X5B2 W5NJ54 A0A3Q3BF84 A0A336MHC5 Q2YDD0 W5U834 A0A3P9PWY1 A0A3P9PWQ1 M3ZS18 L8INA2 A0A315V9H2 A0A3B3U5B4 A0A3B3VHG6 A0A1A8PB39 A0A1A8LVI3 A0A2I4BQK9 A0A2Y9KCJ1 A0A3Q7TFY1 A0A3Q7T552 A0A1A8A8L2 A0A3Q1H6G9 A0A2U3V2E4 E2RHI0 A0A2U3WS28 A0A3Q1C0N7 A0A3P8TDC0 A0A1A8V9N8 A0A1A8BE07 A0A3B5PXX6 A0A3Q2LPT5 A0A340XLE4 F7AUE0 A0A341AUR3 W5KQY9 A0A2Y9NYU7 A0A3Q0RC56 A0A2Y9NTG0 A0A1S3AAR9 A0A2Y9FLC5 A0A384B9Z4 A0A1A8JIH6 A0A1A8FRG3 A0A1A8J2G4 A0A2Y9HL33 M3WAW7 A0A1A8FGW6 A0A1A7YF47 A0A146MM25 I3K9M5 A0A3B4DNV6 A0A3Q7MQ43 G3SY46 A0A3Q2Z7K7 A0A2U3XV83 F7IA97 F1RTY7 A0A2K5D5F4 U3CHH0 U3BDK8 G1LYF4 A0A3B4ZE85 F6VN33 A0A3B4ZE58 A0A3Q3JVW0 G2HEZ2 A0A1S3F0L6 A0A3B3QW90 H2N9T6 A0A2U9BAY7 A0A2R8ZHF6 L5LUB0 A0A2J8VKL9 A0A3P8PW12
PDB
1S3X
E-value=8.61999e-48,
Score=481
Ontologies
GO
PANTHER
Topology
Subcellular location
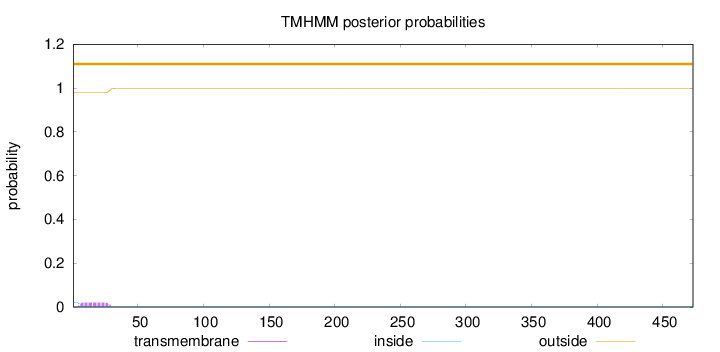
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.454130000000001
Exp number, first 60 AAs:
0.4535
Total prob of N-in:
0.02118
outside
1 - 473
Population Genetic Test Statistics
Pi
189.006342
Theta
180.706385
Tajima's D
0.144513
CLR
0.001983
CSRT
0.411979401029949
Interpretation
Uncertain
