Gene
KWMTBOMO05596
Annotation
PREDICTED:_multiple_inositol_polyphosphate_phosphatase_1_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.92 Nuclear Reliability : 1.528
Sequence
CDS
ATGGTAATAAGACATGGAACAAGATATCCTAATGCTAAGGATATAACAAAAATAAATACACGATTAAAAGAGTTGAAGCTGGAGATATTACTGAGACATAGGCTTGGCAAAGGTGAGCTATCCGATGAACAAATAAAAAAAATCGAAAGTTGGACATGGGACTTGGATTTAGATAAAGAAAAATTCTTAACATTAGAGGGACAAGATGAAATGATAATTTTAGCAGAAAGAACACGAAAACGATTTCCGGGAGCTGTTAAAGAAAAATATGACAATCAGACATTCCTGTTTCGCTACACGGCAACACAAAGAGCACAACAAAGCGCAAGATATTTTGCAAATGGGCTCTTCGACAAGAAGGAGGCCCAAGATGTAATATTCTCTCCGGCGGCCAAAGTTGACACAGTTTTAAGGTTTTACAAACACTGTGACAAGTGGCAAAAGCAAGTAAAAAAGAATCCATTAACATACACGGAACAAAAACTCTATGGGATTAGCAAAGAAATGAATGACACAATTAAATCGGTCACGAAAAGGCTTGATGACGTGAACTTAATGTACAAAGCTTGCGGCTTTGAAACTTCATGGAATAAACATTTCACATCGCCCTGGTGTTACCCTTTTGATGTCAAAAGTGTTCAGACAATGGAATATTATCAGGATTTGAAATATTATTGGATGGACGGATACGGTCACGATTTGACGTACCGCCAGGCTTGCGTCTCCATCAAGAATATGTTTGAGAATTTTAGTTCAAAAGGGGGTCCCAGCGCTTCATTCCTGTTCACACATTCCGGAACGATATTAAAAATATTAACGCATTTGGAACTTTTCAAGCCACGTTCGCACTTGAGAGGCGACAAAAACGTACCGGACAGAACTTGGAGGGCTTCTGACATCGATTGTTTCGGAGCTAATCTAGCGCTGGTTTTGTACAGATGTAAAGGTGTAGACTACGTGCTTGCGTTGCATCAAGAGCGCGTCGTCAAACTGCCGATGTGCGAAAAAGAGCTTTGTCCATTAAAAACGCTAAAGGAATATTTTCATAAATCGATTTACGAATGCGATTACACCGACATGTGCAGTTTGAAGAAGGACGATCTATAG
Protein
MVIRHGTRYPNAKDITKINTRLKELKLEILLRHRLGKGELSDEQIKKIESWTWDLDLDKEKFLTLEGQDEMIILAERTRKRFPGAVKEKYDNQTFLFRYTATQRAQQSARYFANGLFDKKEAQDVIFSPAAKVDTVLRFYKHCDKWQKQVKKNPLTYTEQKLYGISKEMNDTIKSVTKRLDDVNLMYKACGFETSWNKHFTSPWCYPFDVKSVQTMEYYQDLKYYWMDGYGHDLTYRQACVSIKNMFENFSSKGGPSASFLFTHSGTILKILTHLELFKPRSHLRGDKNVPDRTWRASDIDCFGANLALVLYRCKGVDYVLALHQERVVKLPMCEKELCPLKTLKEYFHKSIYECDYTDMCSLKKDDL
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
A0A2A4JZC4
A0A194PFX8
A0A212FKE5
A0A1C9EGM1
H9JAN0
E0VP92
+ More
A0A2J7PTP3 N6T0V9 A0A067RMS2 A0A182HZ88 Q7PQI2 A0A1B6GP03 A0A182JSJ0 A0A182WVV0 A0A182RZE6 A0A084VB11 A0A182KXF7 A0A182PH50 A0A1L8DZL8 A0A1B6D218 A0A182VS01 A0A0C9QVI9 A0A182NSN5 A0A0N0BC56 A0A1L8DZM9 A0A1L8E054 A0A0L7R8L9 A0A182Y7F2 A0A2M4ATC0 W5JWY6 A0A034VSL6 A0A182F715 A0A2M4ASX3 A0A182PZT2 A0A2M4BM93 A0A2M4BN15 T1DNS6 A0A2M4BM50 A0A2S2QWD5 A0A0A1WV45 A0A0T6BFU1 A0A154PEM8 A0A1I8PYE6 A0A1B6LLB7 A0A2A3E3A8 W8BG64 A0A088A6I1 A0A1W4WIW2 A0A2H8TS57 B4L6G5 A0A1B0DG89 A0A1B6E9P5 B4JNN0 E9G9W3 A0A1Y1LXL9 A0A1I8NG08 A0A182TIX3 A0A182V0R0 B4M841 A0A1L8DZM6 A0A182S7F2 E9J8X1 U5EQ88 A0A0Q5T5E6 A0A0P8YKL3 A0A182GZS4 A0A1B0CJK3 A0A026VZI3 J9K731 Q0IEB0 A0A158NJ92 F4WKQ5 A0A151JM62 A0A151JZ28 A0A1W4VSQ6 A0A182MAV2 B4I0F0 Q9W438 O96420 A0A0P5ZU34 A0A0R1E9N5 A0A1S4FT61 A0A0N8D4B2 A0A195C2W3 A0A0P4ZC10 A0A0P5U8D8 A0A0A9XX43 A0A0P4YM59 A0A0P5TJ16 A0A0R3P0A2 A0A0P5AVM8 E2AXD1 E9G9W2 A0A0P5BKZ3 E2BSX4 A0A164X172 A0A0P4Y6F2 A0A0P6IXT1 A0A195BJL0 A0A0P4YQH5
A0A2J7PTP3 N6T0V9 A0A067RMS2 A0A182HZ88 Q7PQI2 A0A1B6GP03 A0A182JSJ0 A0A182WVV0 A0A182RZE6 A0A084VB11 A0A182KXF7 A0A182PH50 A0A1L8DZL8 A0A1B6D218 A0A182VS01 A0A0C9QVI9 A0A182NSN5 A0A0N0BC56 A0A1L8DZM9 A0A1L8E054 A0A0L7R8L9 A0A182Y7F2 A0A2M4ATC0 W5JWY6 A0A034VSL6 A0A182F715 A0A2M4ASX3 A0A182PZT2 A0A2M4BM93 A0A2M4BN15 T1DNS6 A0A2M4BM50 A0A2S2QWD5 A0A0A1WV45 A0A0T6BFU1 A0A154PEM8 A0A1I8PYE6 A0A1B6LLB7 A0A2A3E3A8 W8BG64 A0A088A6I1 A0A1W4WIW2 A0A2H8TS57 B4L6G5 A0A1B0DG89 A0A1B6E9P5 B4JNN0 E9G9W3 A0A1Y1LXL9 A0A1I8NG08 A0A182TIX3 A0A182V0R0 B4M841 A0A1L8DZM6 A0A182S7F2 E9J8X1 U5EQ88 A0A0Q5T5E6 A0A0P8YKL3 A0A182GZS4 A0A1B0CJK3 A0A026VZI3 J9K731 Q0IEB0 A0A158NJ92 F4WKQ5 A0A151JM62 A0A151JZ28 A0A1W4VSQ6 A0A182MAV2 B4I0F0 Q9W438 O96420 A0A0P5ZU34 A0A0R1E9N5 A0A1S4FT61 A0A0N8D4B2 A0A195C2W3 A0A0P4ZC10 A0A0P5U8D8 A0A0A9XX43 A0A0P4YM59 A0A0P5TJ16 A0A0R3P0A2 A0A0P5AVM8 E2AXD1 E9G9W2 A0A0P5BKZ3 E2BSX4 A0A164X172 A0A0P4Y6F2 A0A0P6IXT1 A0A195BJL0 A0A0P4YQH5
Pubmed
26354079
22118469
19121390
20566863
23537049
24845553
+ More
12364791 14747013 17210077 24438588 20966253 25244985 20920257 23761445 25348373 25830018 24495485 17994087 21292972 28004739 25315136 21282665 26483478 24508170 17510324 21347285 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10087200 17550304 25401762 26823975 15632085 20798317
12364791 14747013 17210077 24438588 20966253 25244985 20920257 23761445 25348373 25830018 24495485 17994087 21292972 28004739 25315136 21282665 26483478 24508170 17510324 21347285 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10087200 17550304 25401762 26823975 15632085 20798317
EMBL
NWSH01000389
PCG76742.1
KQ459606
KPI91604.1
AGBW02008063
OWR54208.1
+ More
KX778610 AON96643.1 BABH01008688 DS235361 EEB15198.1 NEVH01021229 PNF19702.1 APGK01056807 KB741277 KB631786 ENN71133.1 ERL86073.1 KK852530 KDR21915.1 APCN01005504 AAAB01008888 EAA08808.5 GECZ01005686 JAS64083.1 ATLV01004843 KE524243 KFB35155.1 GFDF01002164 JAV11920.1 GEDC01017645 JAS19653.1 GBYB01000074 GBYB01007729 JAG69841.1 JAG77496.1 KQ435922 KOX68518.1 GFDF01002161 JAV11923.1 GFDF01002162 JAV11922.1 KQ414632 KOC67227.1 GGFK01010641 MBW43962.1 ADMH02000081 ETN67875.1 GAKP01014152 JAC44800.1 GGFK01010558 MBW43879.1 AXCN02000233 GGFJ01004920 MBW54061.1 GGFJ01005338 MBW54479.1 GAMD01002610 JAA98980.1 GGFJ01004933 MBW54074.1 GGMS01012842 MBY82045.1 GBXI01011358 JAD02934.1 LJIG01000730 KRT86210.1 KQ434874 KZC09738.1 GEBQ01015467 JAT24510.1 KZ288405 PBC26190.1 GAMC01006265 JAC00291.1 GFXV01005025 MBW16830.1 CH933812 EDW05961.1 AJVK01059410 GEDC01002640 JAS34658.1 CH916371 EDV92323.1 GL732536 EFX83809.1 GEZM01044418 JAV78239.1 CH940653 EDW62317.2 GFDF01002163 JAV11921.1 GL769162 EFZ10731.1 GANO01003377 JAB56494.1 CH954180 KQS30450.1 CH902621 KPU81609.1 JXUM01002151 AJWK01014690 AJWK01014691 KK107536 EZA49193.1 ABLF02030102 CH477764 EAT36504.1 ADTU01017592 GL888206 EGI65147.1 KQ978949 KYN27390.1 KQ981427 KYN41772.1 AXCM01001479 CH480819 EDW52981.1 AE014298 AY095178 AAF46121.1 AAM12271.1 AF046912 AAD02435.1 GDIP01052105 JAM51610.1 CM000162 KRK05855.1 GDIP01070151 JAM33564.1 KQ978317 KYM95177.1 GDIP01216140 GDIP01092605 GDIP01043270 JAJ07262.1 GDIP01118197 JAL85517.1 GBHO01021759 GBHO01021758 GBRD01011509 GDHC01017255 GDHC01001896 JAG21845.1 JAG21846.1 JAG54315.1 JAQ01374.1 JAQ16733.1 GDIP01226840 JAI96561.1 GDIP01127154 JAL76560.1 CH379064 KRT06650.1 GDIP01198047 JAJ25355.1 GL443548 EFN61911.1 EFX83810.1 GDIP01198048 JAJ25354.1 GL450311 EFN81199.1 LRGB01001036 KZS13773.1 GDIP01232899 JAI90502.1 GDIQ01026114 JAN68623.1 KQ976453 KYM85376.1 GDIP01224285 GDIP01124790 JAI99116.1
KX778610 AON96643.1 BABH01008688 DS235361 EEB15198.1 NEVH01021229 PNF19702.1 APGK01056807 KB741277 KB631786 ENN71133.1 ERL86073.1 KK852530 KDR21915.1 APCN01005504 AAAB01008888 EAA08808.5 GECZ01005686 JAS64083.1 ATLV01004843 KE524243 KFB35155.1 GFDF01002164 JAV11920.1 GEDC01017645 JAS19653.1 GBYB01000074 GBYB01007729 JAG69841.1 JAG77496.1 KQ435922 KOX68518.1 GFDF01002161 JAV11923.1 GFDF01002162 JAV11922.1 KQ414632 KOC67227.1 GGFK01010641 MBW43962.1 ADMH02000081 ETN67875.1 GAKP01014152 JAC44800.1 GGFK01010558 MBW43879.1 AXCN02000233 GGFJ01004920 MBW54061.1 GGFJ01005338 MBW54479.1 GAMD01002610 JAA98980.1 GGFJ01004933 MBW54074.1 GGMS01012842 MBY82045.1 GBXI01011358 JAD02934.1 LJIG01000730 KRT86210.1 KQ434874 KZC09738.1 GEBQ01015467 JAT24510.1 KZ288405 PBC26190.1 GAMC01006265 JAC00291.1 GFXV01005025 MBW16830.1 CH933812 EDW05961.1 AJVK01059410 GEDC01002640 JAS34658.1 CH916371 EDV92323.1 GL732536 EFX83809.1 GEZM01044418 JAV78239.1 CH940653 EDW62317.2 GFDF01002163 JAV11921.1 GL769162 EFZ10731.1 GANO01003377 JAB56494.1 CH954180 KQS30450.1 CH902621 KPU81609.1 JXUM01002151 AJWK01014690 AJWK01014691 KK107536 EZA49193.1 ABLF02030102 CH477764 EAT36504.1 ADTU01017592 GL888206 EGI65147.1 KQ978949 KYN27390.1 KQ981427 KYN41772.1 AXCM01001479 CH480819 EDW52981.1 AE014298 AY095178 AAF46121.1 AAM12271.1 AF046912 AAD02435.1 GDIP01052105 JAM51610.1 CM000162 KRK05855.1 GDIP01070151 JAM33564.1 KQ978317 KYM95177.1 GDIP01216140 GDIP01092605 GDIP01043270 JAJ07262.1 GDIP01118197 JAL85517.1 GBHO01021759 GBHO01021758 GBRD01011509 GDHC01017255 GDHC01001896 JAG21845.1 JAG21846.1 JAG54315.1 JAQ01374.1 JAQ16733.1 GDIP01226840 JAI96561.1 GDIP01127154 JAL76560.1 CH379064 KRT06650.1 GDIP01198047 JAJ25355.1 GL443548 EFN61911.1 EFX83810.1 GDIP01198048 JAJ25354.1 GL450311 EFN81199.1 LRGB01001036 KZS13773.1 GDIP01232899 JAI90502.1 GDIQ01026114 JAN68623.1 KQ976453 KYM85376.1 GDIP01224285 GDIP01124790 JAI99116.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000005204
UP000009046
UP000235965
+ More
UP000019118 UP000030742 UP000027135 UP000075840 UP000007062 UP000075881 UP000076407 UP000075900 UP000030765 UP000075882 UP000075885 UP000075920 UP000075884 UP000053105 UP000053825 UP000076408 UP000000673 UP000069272 UP000075886 UP000076502 UP000095300 UP000242457 UP000005203 UP000192223 UP000009192 UP000092462 UP000001070 UP000000305 UP000095301 UP000075902 UP000075903 UP000008792 UP000075901 UP000008711 UP000007801 UP000069940 UP000092461 UP000053097 UP000007819 UP000008820 UP000005205 UP000007755 UP000078492 UP000078541 UP000192221 UP000075883 UP000001292 UP000000803 UP000002282 UP000078542 UP000001819 UP000000311 UP000008237 UP000076858 UP000078540
UP000019118 UP000030742 UP000027135 UP000075840 UP000007062 UP000075881 UP000076407 UP000075900 UP000030765 UP000075882 UP000075885 UP000075920 UP000075884 UP000053105 UP000053825 UP000076408 UP000000673 UP000069272 UP000075886 UP000076502 UP000095300 UP000242457 UP000005203 UP000192223 UP000009192 UP000092462 UP000001070 UP000000305 UP000095301 UP000075902 UP000075903 UP000008792 UP000075901 UP000008711 UP000007801 UP000069940 UP000092461 UP000053097 UP000007819 UP000008820 UP000005205 UP000007755 UP000078492 UP000078541 UP000192221 UP000075883 UP000001292 UP000000803 UP000002282 UP000078542 UP000001819 UP000000311 UP000008237 UP000076858 UP000078540
Interpro
SUPFAM
SSF53254
SSF53254
Gene 3D
ProteinModelPortal
A0A2A4JZC4
A0A194PFX8
A0A212FKE5
A0A1C9EGM1
H9JAN0
E0VP92
+ More
A0A2J7PTP3 N6T0V9 A0A067RMS2 A0A182HZ88 Q7PQI2 A0A1B6GP03 A0A182JSJ0 A0A182WVV0 A0A182RZE6 A0A084VB11 A0A182KXF7 A0A182PH50 A0A1L8DZL8 A0A1B6D218 A0A182VS01 A0A0C9QVI9 A0A182NSN5 A0A0N0BC56 A0A1L8DZM9 A0A1L8E054 A0A0L7R8L9 A0A182Y7F2 A0A2M4ATC0 W5JWY6 A0A034VSL6 A0A182F715 A0A2M4ASX3 A0A182PZT2 A0A2M4BM93 A0A2M4BN15 T1DNS6 A0A2M4BM50 A0A2S2QWD5 A0A0A1WV45 A0A0T6BFU1 A0A154PEM8 A0A1I8PYE6 A0A1B6LLB7 A0A2A3E3A8 W8BG64 A0A088A6I1 A0A1W4WIW2 A0A2H8TS57 B4L6G5 A0A1B0DG89 A0A1B6E9P5 B4JNN0 E9G9W3 A0A1Y1LXL9 A0A1I8NG08 A0A182TIX3 A0A182V0R0 B4M841 A0A1L8DZM6 A0A182S7F2 E9J8X1 U5EQ88 A0A0Q5T5E6 A0A0P8YKL3 A0A182GZS4 A0A1B0CJK3 A0A026VZI3 J9K731 Q0IEB0 A0A158NJ92 F4WKQ5 A0A151JM62 A0A151JZ28 A0A1W4VSQ6 A0A182MAV2 B4I0F0 Q9W438 O96420 A0A0P5ZU34 A0A0R1E9N5 A0A1S4FT61 A0A0N8D4B2 A0A195C2W3 A0A0P4ZC10 A0A0P5U8D8 A0A0A9XX43 A0A0P4YM59 A0A0P5TJ16 A0A0R3P0A2 A0A0P5AVM8 E2AXD1 E9G9W2 A0A0P5BKZ3 E2BSX4 A0A164X172 A0A0P4Y6F2 A0A0P6IXT1 A0A195BJL0 A0A0P4YQH5
A0A2J7PTP3 N6T0V9 A0A067RMS2 A0A182HZ88 Q7PQI2 A0A1B6GP03 A0A182JSJ0 A0A182WVV0 A0A182RZE6 A0A084VB11 A0A182KXF7 A0A182PH50 A0A1L8DZL8 A0A1B6D218 A0A182VS01 A0A0C9QVI9 A0A182NSN5 A0A0N0BC56 A0A1L8DZM9 A0A1L8E054 A0A0L7R8L9 A0A182Y7F2 A0A2M4ATC0 W5JWY6 A0A034VSL6 A0A182F715 A0A2M4ASX3 A0A182PZT2 A0A2M4BM93 A0A2M4BN15 T1DNS6 A0A2M4BM50 A0A2S2QWD5 A0A0A1WV45 A0A0T6BFU1 A0A154PEM8 A0A1I8PYE6 A0A1B6LLB7 A0A2A3E3A8 W8BG64 A0A088A6I1 A0A1W4WIW2 A0A2H8TS57 B4L6G5 A0A1B0DG89 A0A1B6E9P5 B4JNN0 E9G9W3 A0A1Y1LXL9 A0A1I8NG08 A0A182TIX3 A0A182V0R0 B4M841 A0A1L8DZM6 A0A182S7F2 E9J8X1 U5EQ88 A0A0Q5T5E6 A0A0P8YKL3 A0A182GZS4 A0A1B0CJK3 A0A026VZI3 J9K731 Q0IEB0 A0A158NJ92 F4WKQ5 A0A151JM62 A0A151JZ28 A0A1W4VSQ6 A0A182MAV2 B4I0F0 Q9W438 O96420 A0A0P5ZU34 A0A0R1E9N5 A0A1S4FT61 A0A0N8D4B2 A0A195C2W3 A0A0P4ZC10 A0A0P5U8D8 A0A0A9XX43 A0A0P4YM59 A0A0P5TJ16 A0A0R3P0A2 A0A0P5AVM8 E2AXD1 E9G9W2 A0A0P5BKZ3 E2BSX4 A0A164X172 A0A0P4Y6F2 A0A0P6IXT1 A0A195BJL0 A0A0P4YQH5
PDB
1QFX
E-value=0.000335705,
Score=104
Ontologies
PATHWAY
GO
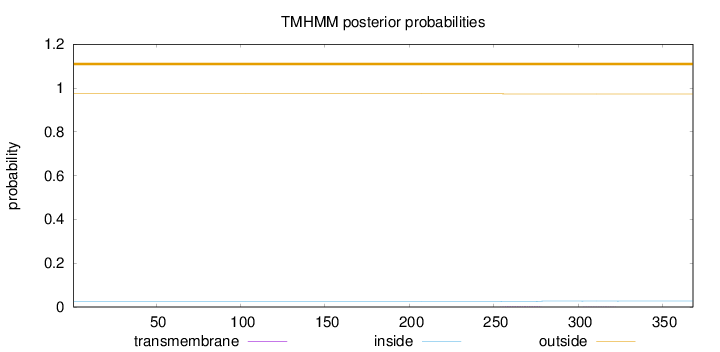
Topology
Length:
368
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04073
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02610
outside
1 - 368
Population Genetic Test Statistics
Pi
240.50456
Theta
160.116815
Tajima's D
1.427078
CLR
0.106505
CSRT
0.773061346932653
Interpretation
Uncertain
