Gene
KWMTBOMO05595
Pre Gene Modal
BGIBMGA006577
Annotation
PREDICTED:_ras-related_protein_Rap-2b_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.381 Nuclear Reliability : 1.115
Sequence
CDS
ATGAAAGGCAGACATCAATTTCGCCGTAGATTCAGTCTGCAGCCTTCCTTCATGAAGGAAGAACACGAAGAAGAGAAGAAACACCACAGGGCTGAAAGAACAGAAGATACAGTACCGAATAGCGCCAATCTCACTAATAACGCAGTGAGGCACAAAATCGTGGTACTAGGTGCGGCTAAAGTCGGCAAATCTTCACTAATCACACAATTCCTATACGGCGCTTTCTCGACCAAATACAAGCGAACCATCGAAGAAATGCATCACGGAGACTTCAATGTATCCGGTGTGAGGTTGACTCTCGACATTTTGGACACGTCCGGATCGTATGAATTCCCGGCAATGCGTACCCTGTCAATGCAATCGGCCGACGCATTCATATTGGTTTACGATATTACGGATGCGAACAGCTTCTCCGAAGTCCGAGCTCTCAGAGACCAGATACACGAAACTAAACAGAGCACGGCCGTTCCTATCGTCGTCGTCGGTAACAAGGTCGATCTGGCAGAGACGGGCGCGCGTCAGGTGGAGTTCCACACAACAGAATCAGTAGTCACAGTCGATTGGGAAAACGGTTTCGTGGAAGCGTCCGCCAAAGACAATATTAACGTATCGCAGATCTTCAAGGAGCTATTGGTACAGGCGAAGGTTAATTTACAGCCGCCTCATATCATCGTTCCTCTAATTCGAGTTGATGTCTAG
Protein
MKGRHQFRRRFSLQPSFMKEEHEEEKKHHRAERTEDTVPNSANLTNNAVRHKIVVLGAAKVGKSSLITQFLYGAFSTKYKRTIEEMHHGDFNVSGVRLTLDILDTSGSYEFPAMRTLSMQSADAFILVYDITDANSFSEVRALRDQIHETKQSTAVPIVVVGNKVDLAETGARQVEFHTTESVVTVDWENGFVEASAKDNINVSQIFKELLVQAKVNLQPPHIIVPLIRVDV
Summary
Uniprot
H9JAN2
A0A194PE36
A0A194QKZ0
A0A1C9EGM8
A0A212F589
A0A3S2LZZ8
+ More
A0A2H1VZK3 A0A2A4JC76 A0A067RDF2 N6T045 D6WG63 A0A182FLJ7 A0A182Y6X4 A0A0L7L7Y8 A0A182QBD2 A0A182LHD6 A0A182RLM4 A0A182HY83 Q7QHQ9 A0A182P0T0 Q17FT2 A0A182JFS8 B4M2I1 B4L4N2 W8BTJ0 Q9W5G7 A0A0M4FBE4 B4R7D1 B4I905 A0A1W4V8Y4 B3P952 B3MSD1 A0A2P8Y5R3 B4PWK0 A0A1I8NMJ4 A0A0Q9X648 Q29HC4 A0A0L0CQ95 A0A3B0K873 A0A1A9W1H4 A0A088AVQ1 A0A1B6LP44 A0A1B0G898 A0A2J7QM89 A0A0M8ZPQ8 B0W2L1 X2JC14 E2B292 A0A026WK08 A0A151JVF2 A0A084VR34 A0A1B6HX15 A0A336MPT2 J9JSE6 E0VP99 F4WKP8 A0A0C9QTN5 A0A2H8TMT7 A0A195BJN9 A0A1S4EC60 A0A151XDL9 T1IEJ6 E2C182 A0A2R7WSN0 A0A0L7RJ47 A0A158NJ85 A0A0J7KJQ5 B4JN71 A0A2A3EQC3 A0A3R7PWA9 A0A182V2V3 A0A2S2NLX2 A0A1I8M6B2 T1ITV7 A0A2J7Q215 A0A151J8Q3 A0A1I8Q5C0 T1ITV6 A0A182TIH9 G3MLH1 A0A2T7PZC1 A0A2P8XMG4 A0A1S3HHK2 A0A1A9X5V2 A0A1B0BSK7 W8BEU6 L7M561 A0A1E1X7P3 A0A023FNR9 G3MM01 A0A023G816 A0A131XGK0 A0A224YP00 A0A131YIJ8 A0A0K8UVS6 A0A2C9JVF8
A0A2H1VZK3 A0A2A4JC76 A0A067RDF2 N6T045 D6WG63 A0A182FLJ7 A0A182Y6X4 A0A0L7L7Y8 A0A182QBD2 A0A182LHD6 A0A182RLM4 A0A182HY83 Q7QHQ9 A0A182P0T0 Q17FT2 A0A182JFS8 B4M2I1 B4L4N2 W8BTJ0 Q9W5G7 A0A0M4FBE4 B4R7D1 B4I905 A0A1W4V8Y4 B3P952 B3MSD1 A0A2P8Y5R3 B4PWK0 A0A1I8NMJ4 A0A0Q9X648 Q29HC4 A0A0L0CQ95 A0A3B0K873 A0A1A9W1H4 A0A088AVQ1 A0A1B6LP44 A0A1B0G898 A0A2J7QM89 A0A0M8ZPQ8 B0W2L1 X2JC14 E2B292 A0A026WK08 A0A151JVF2 A0A084VR34 A0A1B6HX15 A0A336MPT2 J9JSE6 E0VP99 F4WKP8 A0A0C9QTN5 A0A2H8TMT7 A0A195BJN9 A0A1S4EC60 A0A151XDL9 T1IEJ6 E2C182 A0A2R7WSN0 A0A0L7RJ47 A0A158NJ85 A0A0J7KJQ5 B4JN71 A0A2A3EQC3 A0A3R7PWA9 A0A182V2V3 A0A2S2NLX2 A0A1I8M6B2 T1ITV7 A0A2J7Q215 A0A151J8Q3 A0A1I8Q5C0 T1ITV6 A0A182TIH9 G3MLH1 A0A2T7PZC1 A0A2P8XMG4 A0A1S3HHK2 A0A1A9X5V2 A0A1B0BSK7 W8BEU6 L7M561 A0A1E1X7P3 A0A023FNR9 G3MM01 A0A023G816 A0A131XGK0 A0A224YP00 A0A131YIJ8 A0A0K8UVS6 A0A2C9JVF8
Pubmed
19121390
26354079
22118469
24845553
23537049
18362917
+ More
19820115 25244985 26227816 20966253 12364791 14747013 17210077 17510324 17994087 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 17550304 15632085 26108605 20798317 24508170 30249741 24438588 20566863 21719571 21347285 25315136 22216098 25576852 28503490 28049606 28797301 26830274 15562597
19820115 25244985 26227816 20966253 12364791 14747013 17210077 17510324 17994087 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 17550304 15632085 26108605 20798317 24508170 30249741 24438588 20566863 21719571 21347285 25315136 22216098 25576852 28503490 28049606 28797301 26830274 15562597
EMBL
BABH01008666
BABH01008667
BABH01008668
KQ459606
KPI91606.1
KQ461198
+ More
KPJ06197.1 KX778610 AON96641.1 AGBW02010242 OWR48869.1 RSAL01000092 RVE47962.1 ODYU01005415 SOQ46223.1 NWSH01002007 PCG69449.1 KK852530 KDR21911.1 APGK01050107 APGK01050108 APGK01050109 APGK01050110 KB741165 KB632353 ENN73429.1 ERL93336.1 KQ971320 EFA00535.2 JTDY01002415 KOB71454.1 AXCN02001110 APCN01004505 AAAB01008816 EAA05193.4 CH477268 EAT45419.1 CH940651 EDW65885.1 CH933810 EDW07510.1 GAMC01004053 JAC02503.1 AE014298 AL050231 AAF45493.2 ACZ95166.1 CAB43324.1 CP012528 ALC49920.1 CM000366 EDX16742.1 CH480825 EDW43686.1 CH954183 EDV45348.1 CH902622 EDV34686.1 PYGN01000897 PSN39581.1 CM000162 EDX00769.1 KRK05782.1 CH964239 KRF99634.1 CH379064 EAL31834.2 JRES01000056 KNC34533.1 OUUW01000016 SPP88862.1 GEBQ01014446 JAT25531.1 CCAG010000214 CCAG010000215 NEVH01013214 PNF29715.1 KQ435922 KOX68407.1 DS231827 EDS29319.1 AHN59206.1 GL445074 EFN60194.1 KK107167 QOIP01000010 EZA56303.1 RLU17497.1 KQ981703 KYN37434.1 ATLV01015442 ATLV01015443 KE525013 KFB40428.1 GECU01028503 JAS79203.1 UFQT01001606 SSX31149.1 ABLF02038456 ABLF02038462 ABLF02038467 ABLF02038470 ABLF02046385 DS235361 EEB15205.1 GL888206 EGI65140.1 GBYB01007084 JAG76851.1 GFXV01002793 MBW14598.1 KQ976453 KYM85388.1 KQ982268 KYQ58457.1 ACPB03015187 GL451891 EFN78303.1 KK855439 PTY22574.1 KQ414582 KOC70839.1 ADTU01017563 LBMM01006603 KMQ90471.1 CH916371 EDV92164.1 KZ288198 PBC33704.1 QCYY01004122 ROT61531.1 GGMR01005323 MBY17942.1 JH431501 NEVH01019374 PNF22623.1 KQ979486 KYN21240.1 JO842722 AEO34339.1 PZQS01000001 PVD38776.1 PYGN01001725 PSN33182.1 JXJN01019755 GAMC01014804 JAB91751.1 GACK01005879 JAA59155.1 GFAC01003891 JAT95297.1 GBBK01001076 JAC23406.1 JO842902 AEO34519.1 GBBM01005481 JAC29937.1 GEFH01003720 JAP64861.1 GFPF01008190 MAA19336.1 GEDV01010245 JAP78312.1 GDHF01021545 JAI30769.1
KPJ06197.1 KX778610 AON96641.1 AGBW02010242 OWR48869.1 RSAL01000092 RVE47962.1 ODYU01005415 SOQ46223.1 NWSH01002007 PCG69449.1 KK852530 KDR21911.1 APGK01050107 APGK01050108 APGK01050109 APGK01050110 KB741165 KB632353 ENN73429.1 ERL93336.1 KQ971320 EFA00535.2 JTDY01002415 KOB71454.1 AXCN02001110 APCN01004505 AAAB01008816 EAA05193.4 CH477268 EAT45419.1 CH940651 EDW65885.1 CH933810 EDW07510.1 GAMC01004053 JAC02503.1 AE014298 AL050231 AAF45493.2 ACZ95166.1 CAB43324.1 CP012528 ALC49920.1 CM000366 EDX16742.1 CH480825 EDW43686.1 CH954183 EDV45348.1 CH902622 EDV34686.1 PYGN01000897 PSN39581.1 CM000162 EDX00769.1 KRK05782.1 CH964239 KRF99634.1 CH379064 EAL31834.2 JRES01000056 KNC34533.1 OUUW01000016 SPP88862.1 GEBQ01014446 JAT25531.1 CCAG010000214 CCAG010000215 NEVH01013214 PNF29715.1 KQ435922 KOX68407.1 DS231827 EDS29319.1 AHN59206.1 GL445074 EFN60194.1 KK107167 QOIP01000010 EZA56303.1 RLU17497.1 KQ981703 KYN37434.1 ATLV01015442 ATLV01015443 KE525013 KFB40428.1 GECU01028503 JAS79203.1 UFQT01001606 SSX31149.1 ABLF02038456 ABLF02038462 ABLF02038467 ABLF02038470 ABLF02046385 DS235361 EEB15205.1 GL888206 EGI65140.1 GBYB01007084 JAG76851.1 GFXV01002793 MBW14598.1 KQ976453 KYM85388.1 KQ982268 KYQ58457.1 ACPB03015187 GL451891 EFN78303.1 KK855439 PTY22574.1 KQ414582 KOC70839.1 ADTU01017563 LBMM01006603 KMQ90471.1 CH916371 EDV92164.1 KZ288198 PBC33704.1 QCYY01004122 ROT61531.1 GGMR01005323 MBY17942.1 JH431501 NEVH01019374 PNF22623.1 KQ979486 KYN21240.1 JO842722 AEO34339.1 PZQS01000001 PVD38776.1 PYGN01001725 PSN33182.1 JXJN01019755 GAMC01014804 JAB91751.1 GACK01005879 JAA59155.1 GFAC01003891 JAT95297.1 GBBK01001076 JAC23406.1 JO842902 AEO34519.1 GBBM01005481 JAC29937.1 GEFH01003720 JAP64861.1 GFPF01008190 MAA19336.1 GEDV01010245 JAP78312.1 GDHF01021545 JAI30769.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000283053
UP000218220
+ More
UP000027135 UP000019118 UP000030742 UP000007266 UP000069272 UP000076408 UP000037510 UP000075886 UP000075882 UP000075900 UP000075840 UP000007062 UP000075885 UP000008820 UP000075880 UP000008792 UP000009192 UP000000803 UP000092553 UP000000304 UP000001292 UP000192221 UP000008711 UP000007801 UP000245037 UP000002282 UP000095300 UP000007798 UP000001819 UP000037069 UP000268350 UP000091820 UP000005203 UP000092444 UP000235965 UP000053105 UP000002320 UP000000311 UP000053097 UP000279307 UP000078541 UP000030765 UP000007819 UP000009046 UP000007755 UP000078540 UP000079169 UP000075809 UP000015103 UP000008237 UP000053825 UP000005205 UP000036403 UP000001070 UP000242457 UP000283509 UP000075903 UP000095301 UP000078492 UP000075902 UP000245119 UP000085678 UP000092443 UP000092460 UP000076420
UP000027135 UP000019118 UP000030742 UP000007266 UP000069272 UP000076408 UP000037510 UP000075886 UP000075882 UP000075900 UP000075840 UP000007062 UP000075885 UP000008820 UP000075880 UP000008792 UP000009192 UP000000803 UP000092553 UP000000304 UP000001292 UP000192221 UP000008711 UP000007801 UP000245037 UP000002282 UP000095300 UP000007798 UP000001819 UP000037069 UP000268350 UP000091820 UP000005203 UP000092444 UP000235965 UP000053105 UP000002320 UP000000311 UP000053097 UP000279307 UP000078541 UP000030765 UP000007819 UP000009046 UP000007755 UP000078540 UP000079169 UP000075809 UP000015103 UP000008237 UP000053825 UP000005205 UP000036403 UP000001070 UP000242457 UP000283509 UP000075903 UP000095301 UP000078492 UP000075902 UP000245119 UP000085678 UP000092443 UP000092460 UP000076420
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JAN2
A0A194PE36
A0A194QKZ0
A0A1C9EGM8
A0A212F589
A0A3S2LZZ8
+ More
A0A2H1VZK3 A0A2A4JC76 A0A067RDF2 N6T045 D6WG63 A0A182FLJ7 A0A182Y6X4 A0A0L7L7Y8 A0A182QBD2 A0A182LHD6 A0A182RLM4 A0A182HY83 Q7QHQ9 A0A182P0T0 Q17FT2 A0A182JFS8 B4M2I1 B4L4N2 W8BTJ0 Q9W5G7 A0A0M4FBE4 B4R7D1 B4I905 A0A1W4V8Y4 B3P952 B3MSD1 A0A2P8Y5R3 B4PWK0 A0A1I8NMJ4 A0A0Q9X648 Q29HC4 A0A0L0CQ95 A0A3B0K873 A0A1A9W1H4 A0A088AVQ1 A0A1B6LP44 A0A1B0G898 A0A2J7QM89 A0A0M8ZPQ8 B0W2L1 X2JC14 E2B292 A0A026WK08 A0A151JVF2 A0A084VR34 A0A1B6HX15 A0A336MPT2 J9JSE6 E0VP99 F4WKP8 A0A0C9QTN5 A0A2H8TMT7 A0A195BJN9 A0A1S4EC60 A0A151XDL9 T1IEJ6 E2C182 A0A2R7WSN0 A0A0L7RJ47 A0A158NJ85 A0A0J7KJQ5 B4JN71 A0A2A3EQC3 A0A3R7PWA9 A0A182V2V3 A0A2S2NLX2 A0A1I8M6B2 T1ITV7 A0A2J7Q215 A0A151J8Q3 A0A1I8Q5C0 T1ITV6 A0A182TIH9 G3MLH1 A0A2T7PZC1 A0A2P8XMG4 A0A1S3HHK2 A0A1A9X5V2 A0A1B0BSK7 W8BEU6 L7M561 A0A1E1X7P3 A0A023FNR9 G3MM01 A0A023G816 A0A131XGK0 A0A224YP00 A0A131YIJ8 A0A0K8UVS6 A0A2C9JVF8
A0A2H1VZK3 A0A2A4JC76 A0A067RDF2 N6T045 D6WG63 A0A182FLJ7 A0A182Y6X4 A0A0L7L7Y8 A0A182QBD2 A0A182LHD6 A0A182RLM4 A0A182HY83 Q7QHQ9 A0A182P0T0 Q17FT2 A0A182JFS8 B4M2I1 B4L4N2 W8BTJ0 Q9W5G7 A0A0M4FBE4 B4R7D1 B4I905 A0A1W4V8Y4 B3P952 B3MSD1 A0A2P8Y5R3 B4PWK0 A0A1I8NMJ4 A0A0Q9X648 Q29HC4 A0A0L0CQ95 A0A3B0K873 A0A1A9W1H4 A0A088AVQ1 A0A1B6LP44 A0A1B0G898 A0A2J7QM89 A0A0M8ZPQ8 B0W2L1 X2JC14 E2B292 A0A026WK08 A0A151JVF2 A0A084VR34 A0A1B6HX15 A0A336MPT2 J9JSE6 E0VP99 F4WKP8 A0A0C9QTN5 A0A2H8TMT7 A0A195BJN9 A0A1S4EC60 A0A151XDL9 T1IEJ6 E2C182 A0A2R7WSN0 A0A0L7RJ47 A0A158NJ85 A0A0J7KJQ5 B4JN71 A0A2A3EQC3 A0A3R7PWA9 A0A182V2V3 A0A2S2NLX2 A0A1I8M6B2 T1ITV7 A0A2J7Q215 A0A151J8Q3 A0A1I8Q5C0 T1ITV6 A0A182TIH9 G3MLH1 A0A2T7PZC1 A0A2P8XMG4 A0A1S3HHK2 A0A1A9X5V2 A0A1B0BSK7 W8BEU6 L7M561 A0A1E1X7P3 A0A023FNR9 G3MM01 A0A023G816 A0A131XGK0 A0A224YP00 A0A131YIJ8 A0A0K8UVS6 A0A2C9JVF8
PDB
4M8N
E-value=2.07647e-27,
Score=301
Ontologies
GO
PANTHER
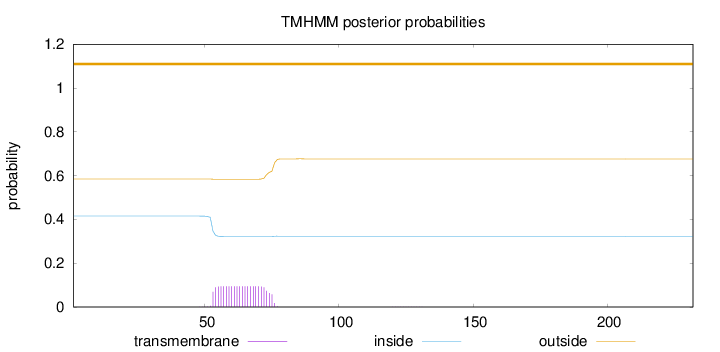
Topology
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.08731
Exp number, first 60 AAs:
0.73483
Total prob of N-in:
0.41525
outside
1 - 232
Population Genetic Test Statistics
Pi
234.923243
Theta
181.601196
Tajima's D
0.639853
CLR
0.795734
CSRT
0.549672516374181
Interpretation
Uncertain
