Pre Gene Modal
BGIBMGA006761
Annotation
PREDICTED:_mitochondrial_inner_membrane_protease_ATP23_homolog_[Bombyx_mori]
Full name
Mitochondrial inner membrane protease ATP23
Location in the cell
Nuclear Reliability : 2.884
Sequence
CDS
ATGTCGGATGGCAGTACTGCTGAGACGAAAACTAACACAGAAAATAATACAAACCAAAAAAATTCTAATGAAAAGAAACCTGATGCATGGGGATACGATTTGTATCCGGAAAGAAAAGGGAGTTTTAAGCCCAAAATAACCAATGTTCTCATTGGAAAAGAAGGAAAAGAGAATATAGAAAAGCTAAAATGTGAACAGAACGTCTTCAAATGTGTACAAACCAGTCCGATCGTTAAAATAATGATGGCTGCTCTCAAAAGTTCAGGCTGTCCCATTGATATCCGACGTCATATATCCTGTGAAGTGTGTGACTACTCAGTCAGTGGTGGTTATGATCCAGAATTGAATCAGATTGTTGTCTGCCAGAATGTATCCACGAGGAAAGGGATGGTACAAGGGGTATTGGCTCATGAAATGATCCACATGTTTGATTTCTGCAGAAACGAATTAGATTTTAAGAACATGGAACATTTGGCTTGTACAGAGATCCGGGCAGCTAATCTCACTCATTGCTCCTTTGCAAGTGCTTGGTCTCAGGGAGATGCATCATGGACTAAGATAAAGAAGGCGCATGAGGATTGTGTGAAGACTAAAGCGCTTTATTCTGTATTGGCTGTACGACAGATAGATAAATTGGAAGCCGTGGACATTATCGAAAAAGTATTTCCTAAGTGCTACCAAGACTTGGAGCCGATCGGCAGGAGGATACGAAGGAATTCCGATGACATGTTCCGAGCTTTCAGAGAAGCCAAATACTATGGTTATGATGATTAG
Protein
MSDGSTAETKTNTENNTNQKNSNEKKPDAWGYDLYPERKGSFKPKITNVLIGKEGKENIEKLKCEQNVFKCVQTSPIVKIMMAALKSSGCPIDIRRHISCEVCDYSVSGGYDPELNQIVVCQNVSTRKGMVQGVLAHEMIHMFDFCRNELDFKNMEHLACTEIRAANLTHCSFASAWSQGDASWTKIKKAHEDCVKTKALYSVLAVRQIDKLEAVDIIEKVFPKCYQDLEPIGRRIRRNSDDMFRAFREAKYYGYDD
Summary
Similarity
Belongs to the peptidase M76 family.
Uniprot
A0A3S2L892
A0A1E1WPT8
A0A194QLF0
A0A194PF09
A0A2A4JD28
A0A1C9EGM0
+ More
A0A2H1WV61 H9JB66 A0A212EVF9 A0A0K8TMA3 A0A336KBE9 U5EVJ1 A0A1Q3F3I0 Q17PI5 A0A1S4EVL9 A0A1B0CX23 B0WC21 A0A182GWX4 A0A0A1WW93 A0A0K8UPE0 A0A034WLB2 T1H6S0 A0A182RI77 A0A1A9UPY0 A0A182KBT8 W8C2I5 A0A182L5W0 A0A1I8NI43 A0A182VG67 A0A182TXD2 Q7Q1Z1 A0A1B0GCS6 A0A2M3ZBD5 A0A182MEK8 A0A2M4AQM8 A0A2M4BWY5 T1DGB7 W5J2X6 A0A023EM18 A0A182Y5P4 B4MYW3 A0A1I8PNY1 A0A1A9WF11 A0A182SJ22 A0A182JAW9 A0A1L8DTE4 A0A1A9Y3U1 A0A1B0BV66 A0A182Q3T6 B4JC87 B3NMG6 B4GQA9 Q29LB4 A0A3B0JH44 B4P8R0 B4NS01 Q9VJD0 B4I5B4 A0A067RQ87 B4LSU3 A0A1J1HUQ7 A0A1W4V010 A0A1W4WFU7 A0A0M4E0J6 B3MMH4 B4KED7 D6WG43 A0A3R7Q0V5 A0A2J7QMC0 A0A1Y1KAT4 A0A0L0CDG3 N6U3J0 U4U5V7 A0A084W3R7 A0A182PB30 A0A182F220 A0A023F309 A0A0N7ZDH6 T1HX28 A0A0N7Z881 A0A0V0GEB6 A0A069DPU0 A0A1B6KM80 A0A0P4YQB6 A0A0N8DA16 E9HK92 A0A182NV02 A0A0P5MS67 A0A182XCL1 A0A182IDY9 A0A1B6FAV4 A0A151JPL4 E2AHV0 F4WZJ5 A0A151X1W4 A0A195BXT8 A0A158NHD1 A0A195ESZ1 A0A0P5F4F6 K7IY35
A0A2H1WV61 H9JB66 A0A212EVF9 A0A0K8TMA3 A0A336KBE9 U5EVJ1 A0A1Q3F3I0 Q17PI5 A0A1S4EVL9 A0A1B0CX23 B0WC21 A0A182GWX4 A0A0A1WW93 A0A0K8UPE0 A0A034WLB2 T1H6S0 A0A182RI77 A0A1A9UPY0 A0A182KBT8 W8C2I5 A0A182L5W0 A0A1I8NI43 A0A182VG67 A0A182TXD2 Q7Q1Z1 A0A1B0GCS6 A0A2M3ZBD5 A0A182MEK8 A0A2M4AQM8 A0A2M4BWY5 T1DGB7 W5J2X6 A0A023EM18 A0A182Y5P4 B4MYW3 A0A1I8PNY1 A0A1A9WF11 A0A182SJ22 A0A182JAW9 A0A1L8DTE4 A0A1A9Y3U1 A0A1B0BV66 A0A182Q3T6 B4JC87 B3NMG6 B4GQA9 Q29LB4 A0A3B0JH44 B4P8R0 B4NS01 Q9VJD0 B4I5B4 A0A067RQ87 B4LSU3 A0A1J1HUQ7 A0A1W4V010 A0A1W4WFU7 A0A0M4E0J6 B3MMH4 B4KED7 D6WG43 A0A3R7Q0V5 A0A2J7QMC0 A0A1Y1KAT4 A0A0L0CDG3 N6U3J0 U4U5V7 A0A084W3R7 A0A182PB30 A0A182F220 A0A023F309 A0A0N7ZDH6 T1HX28 A0A0N7Z881 A0A0V0GEB6 A0A069DPU0 A0A1B6KM80 A0A0P4YQB6 A0A0N8DA16 E9HK92 A0A182NV02 A0A0P5MS67 A0A182XCL1 A0A182IDY9 A0A1B6FAV4 A0A151JPL4 E2AHV0 F4WZJ5 A0A151X1W4 A0A195BXT8 A0A158NHD1 A0A195ESZ1 A0A0P5F4F6 K7IY35
EC Number
3.4.24.-
Pubmed
26354079
19121390
22118469
26369729
17510324
26483478
+ More
25830018 25348373 24495485 20966253 25315136 12364791 14747013 17210077 20920257 23761445 24945155 25244985 17994087 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 18362917 19820115 28004739 26108605 23537049 24438588 25474469 27129103 26334808 21292972 20798317 21719571 21347285 20075255
25830018 25348373 24495485 20966253 25315136 12364791 14747013 17210077 20920257 23761445 24945155 25244985 17994087 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 18362917 19820115 28004739 26108605 23537049 24438588 25474469 27129103 26334808 21292972 20798317 21719571 21347285 20075255
EMBL
RSAL01000092
RVE47964.1
GDQN01002019
JAT89035.1
KQ461198
KPJ06199.1
+ More
KQ459606 KPI91608.1 NWSH01002007 PCG69454.1 KX778610 AON96639.1 ODYU01011300 SOQ56948.1 BABH01008658 BABH01008659 AGBW02012210 OWR45447.1 GDAI01002119 JAI15484.1 UFQS01000236 UFQT01000236 SSX01811.1 SSX22188.1 GANO01003383 JAB56488.1 GFDL01012958 JAV22087.1 CH477191 EAT48632.1 AJWK01033138 DS231884 EDS43081.1 JXUM01094271 KQ564117 KXJ72748.1 GBXI01010998 JAD03294.1 GDHF01031809 GDHF01028605 GDHF01023762 GDHF01010425 GDHF01009437 GDHF01006475 JAI20505.1 JAI23709.1 JAI28552.1 JAI41889.1 JAI42877.1 JAI45839.1 GAKP01003508 JAC55444.1 GAMC01000433 JAC06123.1 AAAB01008980 EAA14499.3 CCAG010002220 GGFM01005106 MBW25857.1 AXCM01001763 GGFK01009760 MBW43081.1 GGFJ01008402 MBW57543.1 GAMD01002831 JAA98759.1 ADMH02002162 ETN58161.1 GAPW01003271 JAC10327.1 CH963913 EDW77302.1 GFDF01004356 JAV09728.1 JXJN01021145 AXCN02000754 CH916368 EDW04120.1 CH954179 EDV54837.1 CH479187 EDW39781.1 CH379060 EAL34131.1 OUUW01000004 SPP80033.1 CM000158 EDW90168.1 CH981725 CM002910 EDX15379.1 KMY90721.1 AE014134 AY118981 AAF53622.2 AAM50841.1 CH480822 EDW55570.1 KK852530 KDR21909.1 CH940649 EDW64854.1 CVRI01000015 CRK89897.1 CP012523 ALC38666.1 CH902620 EDV30920.1 CH933807 EDW12905.1 KQ971319 EFA00208.1 QCYY01003055 ROT65689.1 NEVH01013214 PNF29717.1 GEZM01087661 JAV58549.1 JRES01000539 KNC30300.1 APGK01050987 KB741180 ENN73132.1 KB631768 ERL85991.1 ATLV01020115 KE525294 KFB44861.1 GBBI01003099 JAC15613.1 GDRN01038005 JAI67272.1 ACPB03015175 GDKW01004083 JAI52512.1 GECL01000445 JAP05679.1 GBGD01002801 JAC86088.1 GEBQ01027441 JAT12536.1 GDIP01226186 JAI97215.1 GDIP01054119 LRGB01001361 JAM49596.1 KZS12576.1 GL732667 EFX67841.1 GDIQ01151985 JAK99740.1 APCN01004540 GECZ01022457 JAS47312.1 KQ978713 KYN29075.1 GL439592 EFN66987.1 GL888479 EGI60439.1 KQ982585 KYQ54389.1 KQ976398 KYM92758.1 ADTU01001636 KQ981979 KYN31370.1 GDIQ01260471 JAJ91253.1
KQ459606 KPI91608.1 NWSH01002007 PCG69454.1 KX778610 AON96639.1 ODYU01011300 SOQ56948.1 BABH01008658 BABH01008659 AGBW02012210 OWR45447.1 GDAI01002119 JAI15484.1 UFQS01000236 UFQT01000236 SSX01811.1 SSX22188.1 GANO01003383 JAB56488.1 GFDL01012958 JAV22087.1 CH477191 EAT48632.1 AJWK01033138 DS231884 EDS43081.1 JXUM01094271 KQ564117 KXJ72748.1 GBXI01010998 JAD03294.1 GDHF01031809 GDHF01028605 GDHF01023762 GDHF01010425 GDHF01009437 GDHF01006475 JAI20505.1 JAI23709.1 JAI28552.1 JAI41889.1 JAI42877.1 JAI45839.1 GAKP01003508 JAC55444.1 GAMC01000433 JAC06123.1 AAAB01008980 EAA14499.3 CCAG010002220 GGFM01005106 MBW25857.1 AXCM01001763 GGFK01009760 MBW43081.1 GGFJ01008402 MBW57543.1 GAMD01002831 JAA98759.1 ADMH02002162 ETN58161.1 GAPW01003271 JAC10327.1 CH963913 EDW77302.1 GFDF01004356 JAV09728.1 JXJN01021145 AXCN02000754 CH916368 EDW04120.1 CH954179 EDV54837.1 CH479187 EDW39781.1 CH379060 EAL34131.1 OUUW01000004 SPP80033.1 CM000158 EDW90168.1 CH981725 CM002910 EDX15379.1 KMY90721.1 AE014134 AY118981 AAF53622.2 AAM50841.1 CH480822 EDW55570.1 KK852530 KDR21909.1 CH940649 EDW64854.1 CVRI01000015 CRK89897.1 CP012523 ALC38666.1 CH902620 EDV30920.1 CH933807 EDW12905.1 KQ971319 EFA00208.1 QCYY01003055 ROT65689.1 NEVH01013214 PNF29717.1 GEZM01087661 JAV58549.1 JRES01000539 KNC30300.1 APGK01050987 KB741180 ENN73132.1 KB631768 ERL85991.1 ATLV01020115 KE525294 KFB44861.1 GBBI01003099 JAC15613.1 GDRN01038005 JAI67272.1 ACPB03015175 GDKW01004083 JAI52512.1 GECL01000445 JAP05679.1 GBGD01002801 JAC86088.1 GEBQ01027441 JAT12536.1 GDIP01226186 JAI97215.1 GDIP01054119 LRGB01001361 JAM49596.1 KZS12576.1 GL732667 EFX67841.1 GDIQ01151985 JAK99740.1 APCN01004540 GECZ01022457 JAS47312.1 KQ978713 KYN29075.1 GL439592 EFN66987.1 GL888479 EGI60439.1 KQ982585 KYQ54389.1 KQ976398 KYM92758.1 ADTU01001636 KQ981979 KYN31370.1 GDIQ01260471 JAJ91253.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000218220
UP000005204
UP000007151
+ More
UP000008820 UP000092461 UP000002320 UP000069940 UP000249989 UP000015102 UP000075900 UP000078200 UP000075881 UP000075882 UP000095301 UP000075903 UP000075902 UP000007062 UP000092444 UP000075883 UP000000673 UP000076408 UP000007798 UP000095300 UP000091820 UP000075901 UP000075880 UP000092443 UP000092460 UP000075886 UP000001070 UP000008711 UP000008744 UP000001819 UP000268350 UP000002282 UP000000304 UP000000803 UP000001292 UP000027135 UP000008792 UP000183832 UP000192221 UP000192223 UP000092553 UP000007801 UP000009192 UP000007266 UP000283509 UP000235965 UP000037069 UP000019118 UP000030742 UP000030765 UP000075885 UP000069272 UP000015103 UP000076858 UP000000305 UP000075884 UP000076407 UP000075840 UP000078492 UP000000311 UP000007755 UP000075809 UP000078540 UP000005205 UP000078541 UP000002358
UP000008820 UP000092461 UP000002320 UP000069940 UP000249989 UP000015102 UP000075900 UP000078200 UP000075881 UP000075882 UP000095301 UP000075903 UP000075902 UP000007062 UP000092444 UP000075883 UP000000673 UP000076408 UP000007798 UP000095300 UP000091820 UP000075901 UP000075880 UP000092443 UP000092460 UP000075886 UP000001070 UP000008711 UP000008744 UP000001819 UP000268350 UP000002282 UP000000304 UP000000803 UP000001292 UP000027135 UP000008792 UP000183832 UP000192221 UP000192223 UP000092553 UP000007801 UP000009192 UP000007266 UP000283509 UP000235965 UP000037069 UP000019118 UP000030742 UP000030765 UP000075885 UP000069272 UP000015103 UP000076858 UP000000305 UP000075884 UP000076407 UP000075840 UP000078492 UP000000311 UP000007755 UP000075809 UP000078540 UP000005205 UP000078541 UP000002358
Pfam
Interpro
IPR019165
Peptidase_M76_ATP23
+ More
IPR019410 Methyltransf_16
IPR029426 FAM86
IPR029063 SAM-dependent_MTases
IPR014720 dsRBD_dom
IPR035699 AAA_6
IPR042228 Dynein_2_C
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR041466 Dynein_AAA5_ext
IPR003593 AAA+_ATPase
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR035706 AAA_9
IPR041589 DNAH3_AAA_lid_1
IPR019410 Methyltransf_16
IPR029426 FAM86
IPR029063 SAM-dependent_MTases
IPR014720 dsRBD_dom
IPR035699 AAA_6
IPR042228 Dynein_2_C
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR041466 Dynein_AAA5_ext
IPR003593 AAA+_ATPase
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR024317 Dynein_heavy_chain_D4_dom
IPR024743 Dynein_HC_stalk
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR035706 AAA_9
IPR041589 DNAH3_AAA_lid_1
Gene 3D
CDD
ProteinModelPortal
A0A3S2L892
A0A1E1WPT8
A0A194QLF0
A0A194PF09
A0A2A4JD28
A0A1C9EGM0
+ More
A0A2H1WV61 H9JB66 A0A212EVF9 A0A0K8TMA3 A0A336KBE9 U5EVJ1 A0A1Q3F3I0 Q17PI5 A0A1S4EVL9 A0A1B0CX23 B0WC21 A0A182GWX4 A0A0A1WW93 A0A0K8UPE0 A0A034WLB2 T1H6S0 A0A182RI77 A0A1A9UPY0 A0A182KBT8 W8C2I5 A0A182L5W0 A0A1I8NI43 A0A182VG67 A0A182TXD2 Q7Q1Z1 A0A1B0GCS6 A0A2M3ZBD5 A0A182MEK8 A0A2M4AQM8 A0A2M4BWY5 T1DGB7 W5J2X6 A0A023EM18 A0A182Y5P4 B4MYW3 A0A1I8PNY1 A0A1A9WF11 A0A182SJ22 A0A182JAW9 A0A1L8DTE4 A0A1A9Y3U1 A0A1B0BV66 A0A182Q3T6 B4JC87 B3NMG6 B4GQA9 Q29LB4 A0A3B0JH44 B4P8R0 B4NS01 Q9VJD0 B4I5B4 A0A067RQ87 B4LSU3 A0A1J1HUQ7 A0A1W4V010 A0A1W4WFU7 A0A0M4E0J6 B3MMH4 B4KED7 D6WG43 A0A3R7Q0V5 A0A2J7QMC0 A0A1Y1KAT4 A0A0L0CDG3 N6U3J0 U4U5V7 A0A084W3R7 A0A182PB30 A0A182F220 A0A023F309 A0A0N7ZDH6 T1HX28 A0A0N7Z881 A0A0V0GEB6 A0A069DPU0 A0A1B6KM80 A0A0P4YQB6 A0A0N8DA16 E9HK92 A0A182NV02 A0A0P5MS67 A0A182XCL1 A0A182IDY9 A0A1B6FAV4 A0A151JPL4 E2AHV0 F4WZJ5 A0A151X1W4 A0A195BXT8 A0A158NHD1 A0A195ESZ1 A0A0P5F4F6 K7IY35
A0A2H1WV61 H9JB66 A0A212EVF9 A0A0K8TMA3 A0A336KBE9 U5EVJ1 A0A1Q3F3I0 Q17PI5 A0A1S4EVL9 A0A1B0CX23 B0WC21 A0A182GWX4 A0A0A1WW93 A0A0K8UPE0 A0A034WLB2 T1H6S0 A0A182RI77 A0A1A9UPY0 A0A182KBT8 W8C2I5 A0A182L5W0 A0A1I8NI43 A0A182VG67 A0A182TXD2 Q7Q1Z1 A0A1B0GCS6 A0A2M3ZBD5 A0A182MEK8 A0A2M4AQM8 A0A2M4BWY5 T1DGB7 W5J2X6 A0A023EM18 A0A182Y5P4 B4MYW3 A0A1I8PNY1 A0A1A9WF11 A0A182SJ22 A0A182JAW9 A0A1L8DTE4 A0A1A9Y3U1 A0A1B0BV66 A0A182Q3T6 B4JC87 B3NMG6 B4GQA9 Q29LB4 A0A3B0JH44 B4P8R0 B4NS01 Q9VJD0 B4I5B4 A0A067RQ87 B4LSU3 A0A1J1HUQ7 A0A1W4V010 A0A1W4WFU7 A0A0M4E0J6 B3MMH4 B4KED7 D6WG43 A0A3R7Q0V5 A0A2J7QMC0 A0A1Y1KAT4 A0A0L0CDG3 N6U3J0 U4U5V7 A0A084W3R7 A0A182PB30 A0A182F220 A0A023F309 A0A0N7ZDH6 T1HX28 A0A0N7Z881 A0A0V0GEB6 A0A069DPU0 A0A1B6KM80 A0A0P4YQB6 A0A0N8DA16 E9HK92 A0A182NV02 A0A0P5MS67 A0A182XCL1 A0A182IDY9 A0A1B6FAV4 A0A151JPL4 E2AHV0 F4WZJ5 A0A151X1W4 A0A195BXT8 A0A158NHD1 A0A195ESZ1 A0A0P5F4F6 K7IY35
Ontologies
GO
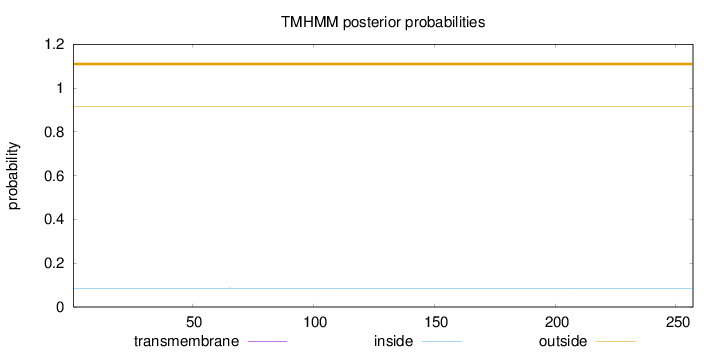
Topology
Length:
257
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00157
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08598
outside
1 - 257
Population Genetic Test Statistics
Pi
217.611357
Theta
175.994881
Tajima's D
0.436031
CLR
0
CSRT
0.488925553722314
Interpretation
Uncertain
