Gene
KWMTBOMO05592
Pre Gene Modal
BGIBMGA006760
Annotation
PREDICTED:_protein-lysine_N-methyltransferase_EEF2KMT_[Papilio_xuthus]
Full name
Protein-lysine N-methyltransferase EEF2KMT
Alternative Name
eEF2-lysine methyltransferase
Location in the cell
Cytoplasmic Reliability : 2.013 Nuclear Reliability : 1.391
Sequence
CDS
ATGAATAACAGTGCTATAGAAGACTATTGTTACCGGCACTATGTTATCAGCAATGATCTTAATAATATTATAACTATGAAAGAAACGAAGAATATGGTTGTTAATGGTACCACAGGAATGCGCACCTGGGAGGCTGCATTGATGCTTTCTGATTGGATTCTATGTAACAAAGAGCTGTTTTCATCAAAAGACGTCCTAGAATTAGGATCAGGTATAGGATTCACTGGTATCACATTAGCAAAATTTTGTGAACCTAAATCAGTAACAATGACTGATTGTCATGAAGATGTTCTACAAGTACTGTGTGAAAATGTGGATATAAATTTTCCATCACAATGTAAAAACAGAAGCTCTGATGGCACAACATATGAATTAGACAATGTTAGTAGAGTTGATGTTAAGATGTTAGATTGGAATGAAATTACCGATCTACCAAATAAAAGGCCCGATGTAATACTTGGTGCAGATATTGTATATGATCCTTCAATACTGAAGCCACTCTGTAATGTACTGAAAATATTCTGTGACAGAAAAAATGACGTGAACATTTATATCGCCAGTATTATAAGAAATGAGTTAACCTTTGAAAGTTTTTTACAAGTTTTGGAAACCTATAATTTTCATTGTGAAAAATTAATGCAACCAACAAATGCGCACATTGAGTGGGATCATACTCATATTAACCGATGCTTACTAAAGATCATGAGGAAATAA
Protein
MNNSAIEDYCYRHYVISNDLNNIITMKETKNMVVNGTTGMRTWEAALMLSDWILCNKELFSSKDVLELGSGIGFTGITLAKFCEPKSVTMTDCHEDVLQVLCENVDINFPSQCKNRSSDGTTYELDNVSRVDVKMLDWNEITDLPNKRPDVILGADIVYDPSILKPLCNVLKIFCDRKNDVNIYIASIIRNELTFESFLQVLETYNFHCEKLMQPTNAHIEWDHTHINRCLLKIMRK
Summary
Description
Catalyzes the trimethylation of eukaryotic elongation factor 2 (EEF2) on 'Lys-525'.
Subunit
Interacts with FAM86B2 and FAM86C1.
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. EEF2KMT family.
Keywords
Acetylation
Alternative splicing
Complete proteome
Cytoplasm
Methyltransferase
Reference proteome
S-adenosyl-L-methionine
Transferase
Feature
chain Protein-lysine N-methyltransferase EEF2KMT
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4JBK2
A0A2H1WV61
A0A212EVC8
A0A3S2NHV3
A0A1C9EGL3
A0A195FN98
+ More
A0A195C637 A0A195AVP7 A0A195E927 A0A0A0AYD6 A0A093RWW6 A0A158NF26 A0A1Q3FFV0 A0A151WHY0 A0A2I4D7V5 A0A1Q3FFM2 A0A1Q3FFQ0 A0A091FL81 A0A1I8QEQ3 R7T7X9 A0A1B6DGU6 B0W680 A0A2D4NBK6 F4WEL4 Q173U5 A0A2I0LWV5 A0A1W4XIQ1 A0A091R895 A0A1V4KT99 A0A182G6Q2 A0A023ERF2 A0A182SX91 A0A226NT43 A0A293N160 A0A0L0C872 A0A087QI92 A0A1W3JTU9 J9JSN5 A0A2D4HVN2 U3I6R4 A0A3B1IIY7 A0A3P8QH70 R0JVP8 H0VPL8 A0A093NZA9 W5L2E3 F6TKJ5 H3D146 I3MQU1 A0A182KC85 A0A091J897 A0A2D4KP71 E9J353 A0A3Q4GQ25 A0A3P9AZX5 A0A093K0A1 A0A091WLE4 A0A182M3V0 A0A182RX58 A0A3Q3CWM6 H3AZ04 A0A182U4P4 A0A182ID75 Q7QB05 Q4SAS7 A0A2M4ARR4 A0A182US78 A0A1V4KT72 H3BW61 A0A182PS79 A0A1A9W1W2 A0A1X7VTR5 A0A1S3ES16 A0A1B6I953 A0A1A6HQA3 T1E9S9 K7GDG6 A0A3B4FQR7 A0A182Y5C1 H3CRR6 A0A2S2PIB1 A0A3Q2PRF2 Q4SN38 A0A093GIC5 F6TPM4 A0A088AJK3 Q3UZW7-2 A0A182FPS2 H0YZ08 A0A182NCY5 A0A1B6IS42 W5J5K2 A0A084WP34 K7GDG1 A0A182W189 A0A2M3Z7V0 A0A2M3Z7R2
A0A195C637 A0A195AVP7 A0A195E927 A0A0A0AYD6 A0A093RWW6 A0A158NF26 A0A1Q3FFV0 A0A151WHY0 A0A2I4D7V5 A0A1Q3FFM2 A0A1Q3FFQ0 A0A091FL81 A0A1I8QEQ3 R7T7X9 A0A1B6DGU6 B0W680 A0A2D4NBK6 F4WEL4 Q173U5 A0A2I0LWV5 A0A1W4XIQ1 A0A091R895 A0A1V4KT99 A0A182G6Q2 A0A023ERF2 A0A182SX91 A0A226NT43 A0A293N160 A0A0L0C872 A0A087QI92 A0A1W3JTU9 J9JSN5 A0A2D4HVN2 U3I6R4 A0A3B1IIY7 A0A3P8QH70 R0JVP8 H0VPL8 A0A093NZA9 W5L2E3 F6TKJ5 H3D146 I3MQU1 A0A182KC85 A0A091J897 A0A2D4KP71 E9J353 A0A3Q4GQ25 A0A3P9AZX5 A0A093K0A1 A0A091WLE4 A0A182M3V0 A0A182RX58 A0A3Q3CWM6 H3AZ04 A0A182U4P4 A0A182ID75 Q7QB05 Q4SAS7 A0A2M4ARR4 A0A182US78 A0A1V4KT72 H3BW61 A0A182PS79 A0A1A9W1W2 A0A1X7VTR5 A0A1S3ES16 A0A1B6I953 A0A1A6HQA3 T1E9S9 K7GDG6 A0A3B4FQR7 A0A182Y5C1 H3CRR6 A0A2S2PIB1 A0A3Q2PRF2 Q4SN38 A0A093GIC5 F6TPM4 A0A088AJK3 Q3UZW7-2 A0A182FPS2 H0YZ08 A0A182NCY5 A0A1B6IS42 W5J5K2 A0A084WP34 K7GDG1 A0A182W189 A0A2M3Z7V0 A0A2M3Z7R2
EC Number
2.1.1.-
Pubmed
EMBL
NWSH01002007
PCG69455.1
ODYU01011300
SOQ56948.1
AGBW02012210
OWR45446.1
+ More
RSAL01000092 RVE47965.1 KX778610 AON96638.1 KQ981490 KYN41364.1 KQ978287 KYM95636.1 KQ976736 KYM76044.1 KQ979479 KYN21334.1 KL873367 KGL98508.1 KL669029 KFW75239.1 ADTU01013712 GFDL01008644 JAV26401.1 KQ983106 KYQ47405.1 GFDL01008710 JAV26335.1 GFDL01008713 JAV26332.1 KL447254 KFO71260.1 AMQN01014733 KB311212 ELT89739.1 GEDC01012463 JAS24835.1 DS231847 EDS36427.1 IACM01154212 LAB42529.1 GL888103 EGI67377.1 CH477416 EAT41351.1 EAT41352.1 AKCR02000070 PKK21915.1 KK810912 KFQ35472.1 LSYS01001700 OPJ87620.1 JXUM01045410 JXUM01063366 KQ562245 KQ561437 KXJ76315.1 KXJ78594.1 GAPW01002349 JAC11249.1 AWGT02000778 OXB70975.1 GFWV01020407 MAA45135.1 JRES01000755 KNC28638.1 KL225618 KFM00946.1 ABLF02027754 IACK01052083 LAA76034.1 KB743068 EOB01626.1 AAKN02007436 AAKN02007437 KL225088 KFW67300.1 AGTP01046858 KL218765 KFP07946.1 IACL01075894 LAB10448.1 GL768057 EFZ12746.1 KL206595 KFV84064.1 KK735903 KFR16452.1 AXCM01000508 AFYH01108017 AFYH01108018 APCN01005045 AAAB01008880 EAA08636.3 CAAE01014679 CAG02255.1 GGFK01010166 MBW43487.1 OPJ87621.1 GECU01024230 JAS83476.1 LZPO01017330 OBS80638.1 GAMD01001052 JAB00539.1 AGCU01096445 GGMR01016007 MBY28626.1 CAAE01014544 CAF97944.1 KL215786 KFV66539.1 AK017533 AK133590 AK163493 AC124576 BC051078 ABQF01051220 GECU01017955 JAS89751.1 ADMH02002178 ETN58044.1 ATLV01024864 KE525362 KFB51978.1 GGFM01003846 MBW24597.1 GGFM01003812 MBW24563.1
RSAL01000092 RVE47965.1 KX778610 AON96638.1 KQ981490 KYN41364.1 KQ978287 KYM95636.1 KQ976736 KYM76044.1 KQ979479 KYN21334.1 KL873367 KGL98508.1 KL669029 KFW75239.1 ADTU01013712 GFDL01008644 JAV26401.1 KQ983106 KYQ47405.1 GFDL01008710 JAV26335.1 GFDL01008713 JAV26332.1 KL447254 KFO71260.1 AMQN01014733 KB311212 ELT89739.1 GEDC01012463 JAS24835.1 DS231847 EDS36427.1 IACM01154212 LAB42529.1 GL888103 EGI67377.1 CH477416 EAT41351.1 EAT41352.1 AKCR02000070 PKK21915.1 KK810912 KFQ35472.1 LSYS01001700 OPJ87620.1 JXUM01045410 JXUM01063366 KQ562245 KQ561437 KXJ76315.1 KXJ78594.1 GAPW01002349 JAC11249.1 AWGT02000778 OXB70975.1 GFWV01020407 MAA45135.1 JRES01000755 KNC28638.1 KL225618 KFM00946.1 ABLF02027754 IACK01052083 LAA76034.1 KB743068 EOB01626.1 AAKN02007436 AAKN02007437 KL225088 KFW67300.1 AGTP01046858 KL218765 KFP07946.1 IACL01075894 LAB10448.1 GL768057 EFZ12746.1 KL206595 KFV84064.1 KK735903 KFR16452.1 AXCM01000508 AFYH01108017 AFYH01108018 APCN01005045 AAAB01008880 EAA08636.3 CAAE01014679 CAG02255.1 GGFK01010166 MBW43487.1 OPJ87621.1 GECU01024230 JAS83476.1 LZPO01017330 OBS80638.1 GAMD01001052 JAB00539.1 AGCU01096445 GGMR01016007 MBY28626.1 CAAE01014544 CAF97944.1 KL215786 KFV66539.1 AK017533 AK133590 AK163493 AC124576 BC051078 ABQF01051220 GECU01017955 JAS89751.1 ADMH02002178 ETN58044.1 ATLV01024864 KE525362 KFB51978.1 GGFM01003846 MBW24597.1 GGFM01003812 MBW24563.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000078541
UP000078542
UP000078540
+ More
UP000078492 UP000053858 UP000053258 UP000005205 UP000075809 UP000192220 UP000053760 UP000095300 UP000014760 UP000002320 UP000007755 UP000008820 UP000053872 UP000192223 UP000190648 UP000069940 UP000249989 UP000075901 UP000198419 UP000037069 UP000053286 UP000007819 UP000018467 UP000265100 UP000005447 UP000054081 UP000002280 UP000007303 UP000005215 UP000075881 UP000054308 UP000261580 UP000265160 UP000053584 UP000053605 UP000075883 UP000075900 UP000264840 UP000008672 UP000075902 UP000075840 UP000007062 UP000075903 UP000075885 UP000091820 UP000007879 UP000081671 UP000092124 UP000007267 UP000261460 UP000076408 UP000265000 UP000053875 UP000008144 UP000005203 UP000000589 UP000069272 UP000007754 UP000075884 UP000000673 UP000030765 UP000075920
UP000078492 UP000053858 UP000053258 UP000005205 UP000075809 UP000192220 UP000053760 UP000095300 UP000014760 UP000002320 UP000007755 UP000008820 UP000053872 UP000192223 UP000190648 UP000069940 UP000249989 UP000075901 UP000198419 UP000037069 UP000053286 UP000007819 UP000018467 UP000265100 UP000005447 UP000054081 UP000002280 UP000007303 UP000005215 UP000075881 UP000054308 UP000261580 UP000265160 UP000053584 UP000053605 UP000075883 UP000075900 UP000264840 UP000008672 UP000075902 UP000075840 UP000007062 UP000075903 UP000075885 UP000091820 UP000007879 UP000081671 UP000092124 UP000007267 UP000261460 UP000076408 UP000265000 UP000053875 UP000008144 UP000005203 UP000000589 UP000069272 UP000007754 UP000075884 UP000000673 UP000030765 UP000075920
PRIDE
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
A0A2A4JBK2
A0A2H1WV61
A0A212EVC8
A0A3S2NHV3
A0A1C9EGL3
A0A195FN98
+ More
A0A195C637 A0A195AVP7 A0A195E927 A0A0A0AYD6 A0A093RWW6 A0A158NF26 A0A1Q3FFV0 A0A151WHY0 A0A2I4D7V5 A0A1Q3FFM2 A0A1Q3FFQ0 A0A091FL81 A0A1I8QEQ3 R7T7X9 A0A1B6DGU6 B0W680 A0A2D4NBK6 F4WEL4 Q173U5 A0A2I0LWV5 A0A1W4XIQ1 A0A091R895 A0A1V4KT99 A0A182G6Q2 A0A023ERF2 A0A182SX91 A0A226NT43 A0A293N160 A0A0L0C872 A0A087QI92 A0A1W3JTU9 J9JSN5 A0A2D4HVN2 U3I6R4 A0A3B1IIY7 A0A3P8QH70 R0JVP8 H0VPL8 A0A093NZA9 W5L2E3 F6TKJ5 H3D146 I3MQU1 A0A182KC85 A0A091J897 A0A2D4KP71 E9J353 A0A3Q4GQ25 A0A3P9AZX5 A0A093K0A1 A0A091WLE4 A0A182M3V0 A0A182RX58 A0A3Q3CWM6 H3AZ04 A0A182U4P4 A0A182ID75 Q7QB05 Q4SAS7 A0A2M4ARR4 A0A182US78 A0A1V4KT72 H3BW61 A0A182PS79 A0A1A9W1W2 A0A1X7VTR5 A0A1S3ES16 A0A1B6I953 A0A1A6HQA3 T1E9S9 K7GDG6 A0A3B4FQR7 A0A182Y5C1 H3CRR6 A0A2S2PIB1 A0A3Q2PRF2 Q4SN38 A0A093GIC5 F6TPM4 A0A088AJK3 Q3UZW7-2 A0A182FPS2 H0YZ08 A0A182NCY5 A0A1B6IS42 W5J5K2 A0A084WP34 K7GDG1 A0A182W189 A0A2M3Z7V0 A0A2M3Z7R2
A0A195C637 A0A195AVP7 A0A195E927 A0A0A0AYD6 A0A093RWW6 A0A158NF26 A0A1Q3FFV0 A0A151WHY0 A0A2I4D7V5 A0A1Q3FFM2 A0A1Q3FFQ0 A0A091FL81 A0A1I8QEQ3 R7T7X9 A0A1B6DGU6 B0W680 A0A2D4NBK6 F4WEL4 Q173U5 A0A2I0LWV5 A0A1W4XIQ1 A0A091R895 A0A1V4KT99 A0A182G6Q2 A0A023ERF2 A0A182SX91 A0A226NT43 A0A293N160 A0A0L0C872 A0A087QI92 A0A1W3JTU9 J9JSN5 A0A2D4HVN2 U3I6R4 A0A3B1IIY7 A0A3P8QH70 R0JVP8 H0VPL8 A0A093NZA9 W5L2E3 F6TKJ5 H3D146 I3MQU1 A0A182KC85 A0A091J897 A0A2D4KP71 E9J353 A0A3Q4GQ25 A0A3P9AZX5 A0A093K0A1 A0A091WLE4 A0A182M3V0 A0A182RX58 A0A3Q3CWM6 H3AZ04 A0A182U4P4 A0A182ID75 Q7QB05 Q4SAS7 A0A2M4ARR4 A0A182US78 A0A1V4KT72 H3BW61 A0A182PS79 A0A1A9W1W2 A0A1X7VTR5 A0A1S3ES16 A0A1B6I953 A0A1A6HQA3 T1E9S9 K7GDG6 A0A3B4FQR7 A0A182Y5C1 H3CRR6 A0A2S2PIB1 A0A3Q2PRF2 Q4SN38 A0A093GIC5 F6TPM4 A0A088AJK3 Q3UZW7-2 A0A182FPS2 H0YZ08 A0A182NCY5 A0A1B6IS42 W5J5K2 A0A084WP34 K7GDG1 A0A182W189 A0A2M3Z7V0 A0A2M3Z7R2
PDB
4QPN
E-value=1.35968e-09,
Score=148
Ontologies
PANTHER
Topology
Subcellular location
Cytoplasm
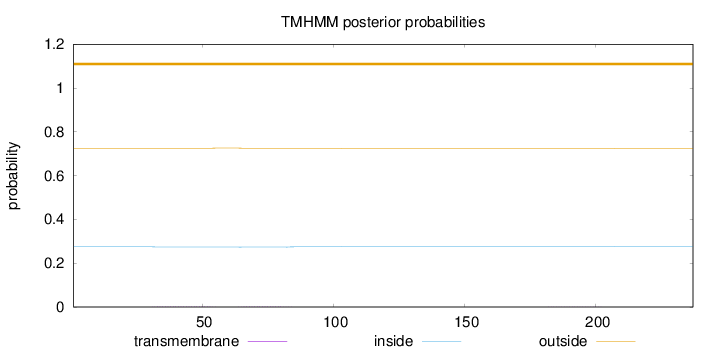
Length:
237
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0658099999999999
Exp number, first 60 AAs:
0.03436
Total prob of N-in:
0.27524
outside
1 - 237
Population Genetic Test Statistics
Pi
320.477064
Theta
188.850486
Tajima's D
2.191875
CLR
0.497323
CSRT
0.910204489775511
Interpretation
Uncertain
