Gene
KWMTBOMO05588
Pre Gene Modal
BGIBMGA006581
Annotation
PREDICTED:_mitogen-activated_protein_kinase_kinase_kinase_kinase_5_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.551
Sequence
CDS
ATGAATTCAGGCACGGACTGGCTCTGCGATATCTTGCAGCATGTACAACTTGAACAATTTTATGTACCAATAAGGGATCAATTACAGATTACCAGACTAGCTCATTTTGATTATGTACATACGGAAGATCTTGAAAGGATTGGTATCAGTAAACCAGGAGTGCGCAGACTATTAGATGCTGTCCGGAAAAAGAAATTGCAGTTATGGAATAGAAAATTTTGGAATAAACTTTTTGGTACATCAAGTTCAGTTGCCAAAGAGAAAACAGATATTGCGAAACCAATTACACAAACTGAAACAAGGACAACATGTATCATACTTGAGAAGGATATTGTGTTACATAATGAACTTGGCAATGGATCATTTGGTGTAGTCAAAAGAGGAGAATGGAAAATTCCAGACCAAACACAAACTGTCCTTGTAGCTGTGAAAGTATTGAAAGCGGATGCATTTGCCCAGCCAGGAATTTATGATGACTTCCGAAGAGAAGTTGAGGCGATGCATAGCTTGAAACATGTGAATTTGATCAAATTACATGGAGTAGTGTTCCATCCATTAATGATGGTCTGCGAACTAGCTCCAATGGGTGCTTTATTAGATTATATAAGAGGTCAAAATGGAAAGGTGTCATTAAATTATATTTCAAAATGGTCAACCCAAGTGGCTGCTGGTATGGCACACCTAGAGAAACACAGATTCCTGCATCGTGATTTAGCTTGTAGAAATATTTTATTGTCAACCTTGGAATTGGTAAAAATTGGTGACTTTGGCCTGATGCGTGCTCTGCCTGATGCGGATGATTGTTATGTCATGAGTGAAAGAAGACGCGTGCCGTTTCCTTGGTGCGCCCCAGAATCTCTGCGCACGAGGCAGTTTAGTCATGCATCTGATGTATGGATGTTTGCTGTTGCCTTGTGGGAAATGTTCACTTTTGGTGAAGAGCCCTGGATAGGCCTCAACGGTTCGGAGATCCTTCGGCTAATAATGCGTGAAGGTCAACGACTTTCGGCCCCCAGTGCGTGTCCCCCGGACGTGTACATGCTCATGATGCAGTGCTGGGACCTGAACCCTAAGGAGAGACCAACGTTCGCCGGCATCCTGCGATACATGGAGACCAACAAATTCGAAACCTCCATAGCATCGTTGAGCTACAGGAAGCAAGGCCAGATGAGTGTGGATGCAGGCGATCCGATTATCCTCATCGACAAAAGACCGGAACTGCACTGGTGGAAGGGGCAAAATCAGAGGTCTTTGGAAGTTGGATTGTTTCCCAGCAACATTCTGTCTGCGGTCAAACTCAAGAACAACCATCCAGCTTCGAAGAAATCCCAAACGTCATCACCCTCTCCTTCGGCAAAATCGTCGGTGCTGAACTCGTCGTTGTCGAATGGAGACGCGCACGAGTCAGCCGTGGTGCTCAGGAAACGCAGAACGATAGAATCGACGCAACCTTCCAGCACGCGCGCCGGTAACGGCGGCAGTAAACAGTTCAACTACAACAAACTTATCAACGATCGCGCTTTGCTGTCCGGCATGCAGAGGTGCACGAGAGACGCCTATTCCAAAAGCTTGAACCAAGTTAAAGAAGACCTGCTAATAGACCTCGATTTGCCGCCTTCGACCAAACCGAGCACACCTAAGCGAGCAACGAACCAAAATAACGTTTCAATAATCGACCAACCAATAGACGTCCCAGAGATTGGCGATGGAATCGAATGGGGCCAGCAAAGCACCGAGAGCTTACCAACGTTCTCAGTGTATTCGAGGAACGCTCAGTCGAGTATGCACGGTGCTAAGTCACTGGACAGTCTCGCGACGTCATCGAACGTGGGCACGTCCACTGGCCAGTGGGCGGAGGATCCCTTCGACGCGTCGTCCTGGCCCGCGGACACCGGCCCGAATGTACACTCGACAAGTTCCCTCAACACCGTCCCAATGCGCCCCAACCACGAACTAGAAGTAAATGAATCAAATTCGATGCGGTCACGACAGAACGAATCTCACGTATACTCGAATAACTACGTCACGACCCGCGACACGGACGCGTCCCGGCTCCACAACATCGCGACCACGTTTAAGAACTCGAATAGTTTAAATACGAATGCACTGATGCAAAATGCGTACAGTTCGTTGGCGGAGATGTCCCTCGACGATAGAATTTCCGAATCGTTGAATCTCAGGTCGAAAACGAATGAAGACAATCTGTACGCGAACAGTACGTACGGCGAGCGACCCGGCACGTCGACCGCGCCCGAGACCCGCTACAAAGACATTCCGATTTATAACAATTTCGATATTAACACCGCCCAACAGCAGTTCATAATCGAAACGAAAGATTACTACACGAAGTTATCGTCTCCGGCGAAAGCGAACACGAGCAACGACTACGAGAAGAACATTTACGTGCCGAAATACGAAGACGAGACGGAGAAACTGAGGAATTTTAGCGACAACGTCGAGAACTCGAAGAATTATTCCGCCTTCAAATATCAGAACGTCGATTACGGTAATTACTCGTACGATTACGGAGCGAATGTCGCTTCGACCTCTCGAGTGCAGTACGACGAGGTCAATGACACTGCCAGTAATTTTTACAGCGAAATAGGCGACGCGTCGAACTTGTACGCGAGCCCTCAGAGAATGTACACAAACGCGTGCTTATATGATGAAGTGTATGAAGAAGCTACGCCTCGTCCGCACCGACCAGCCCCGCCTTGTCCAAAGAAACAAAAATAA
Protein
MNSGTDWLCDILQHVQLEQFYVPIRDQLQITRLAHFDYVHTEDLERIGISKPGVRRLLDAVRKKKLQLWNRKFWNKLFGTSSSVAKEKTDIAKPITQTETRTTCIILEKDIVLHNELGNGSFGVVKRGEWKIPDQTQTVLVAVKVLKADAFAQPGIYDDFRREVEAMHSLKHVNLIKLHGVVFHPLMMVCELAPMGALLDYIRGQNGKVSLNYISKWSTQVAAGMAHLEKHRFLHRDLACRNILLSTLELVKIGDFGLMRALPDADDCYVMSERRRVPFPWCAPESLRTRQFSHASDVWMFAVALWEMFTFGEEPWIGLNGSEILRLIMREGQRLSAPSACPPDVYMLMMQCWDLNPKERPTFAGILRYMETNKFETSIASLSYRKQGQMSVDAGDPIILIDKRPELHWWKGQNQRSLEVGLFPSNILSAVKLKNNHPASKKSQTSSPSPSAKSSVLNSSLSNGDAHESAVVLRKRRTIESTQPSSTRAGNGGSKQFNYNKLINDRALLSGMQRCTRDAYSKSLNQVKEDLLIDLDLPPSTKPSTPKRATNQNNVSIIDQPIDVPEIGDGIEWGQQSTESLPTFSVYSRNAQSSMHGAKSLDSLATSSNVGTSTGQWAEDPFDASSWPADTGPNVHSTSSLNTVPMRPNHELEVNESNSMRSRQNESHVYSNNYVTTRDTDASRLHNIATTFKNSNSLNTNALMQNAYSSLAEMSLDDRISESLNLRSKTNEDNLYANSTYGERPGTSTAPETRYKDIPIYNNFDINTAQQQFIIETKDYYTKLSSPAKANTSNDYEKNIYVPKYEDETEKLRNFSDNVENSKNYSAFKYQNVDYGNYSYDYGANVASTSRVQYDEVNDTASNFYSEIGDASNLYASPQRMYTNACLYDEVYEEATPRPHRPAPPCPKKQK
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
EMBL
BABH01008651
BABH01008652
BABH01008653
BABH01008654
NWSH01000874
PCG73771.1
+ More
RSAL01000092 RVE47968.1 KQ461198 KPJ06202.1 ODYU01003265 SOQ41793.1 KQ459606 KPI91612.1 KX778610 AON96635.1 GDHF01022677 GDHF01004163 JAI29637.1 JAI48151.1 OUUW01000002 SPP76785.1 CH379070 EAL30528.2 GEDC01022770 JAS14528.1
RSAL01000092 RVE47968.1 KQ461198 KPJ06202.1 ODYU01003265 SOQ41793.1 KQ459606 KPI91612.1 KX778610 AON96635.1 GDHF01022677 GDHF01004163 JAI29637.1 JAI48151.1 OUUW01000002 SPP76785.1 CH379070 EAL30528.2 GEDC01022770 JAS14528.1
Proteomes
PRIDE
Interpro
IPR001245
Ser-Thr/Tyr_kinase_cat_dom
+ More
IPR017441 Protein_kinase_ATP_BS
IPR008266 Tyr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR036028 SH3-like_dom_sf
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001452 SH3_domain
IPR015940 UBA
IPR037085 Cdc42-bd-like_dom_sf
IPR015116 Cdc42_binding_dom-like
IPR017441 Protein_kinase_ATP_BS
IPR008266 Tyr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR036028 SH3-like_dom_sf
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001452 SH3_domain
IPR015940 UBA
IPR037085 Cdc42-bd-like_dom_sf
IPR015116 Cdc42_binding_dom-like
Gene 3D
ProteinModelPortal
PDB
4HZS
E-value=1.26712e-102,
Score=957
Ontologies
GO
Topology
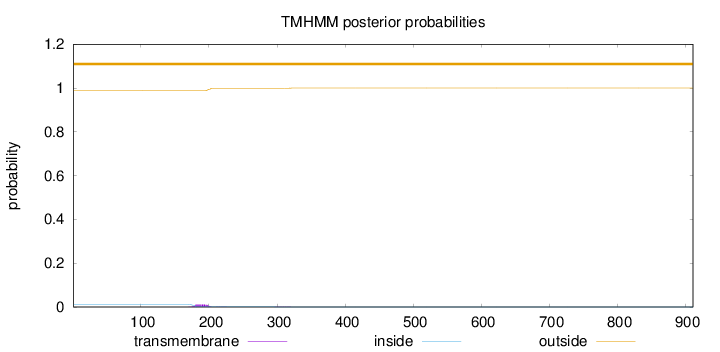
Length:
911
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25991
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01122
outside
1 - 911
Population Genetic Test Statistics
Pi
312.804397
Theta
183.800621
Tajima's D
2.237239
CLR
0.237191
CSRT
0.916354182290885
Interpretation
Uncertain
