Gene
KWMTBOMO05587
Pre Gene Modal
BGIBMGA006582
Annotation
PREDICTED:_receptor-type_tyrosine-protein_phosphatase_N2_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.349 Nuclear Reliability : 1.655
Sequence
CDS
ATGTACGGCCTGTCTTTGCGGCAGGCTCTGTGGGCGCTGGCGTTGATAAACGCGCTGGCGCCCTCGCACGCCGATGGTAACATAGGTTGCTTGTTCAGCGCACCACTGTGTGTAGAGGGCGTAGAGTGGTGCTACGATGATTTCGCCTTTGGGAAATGCATTCCGGTTTACGAGGAAGAGTCAGAAGAAGGCGCGCTCTACCGTTACGACATGGGTCCGGAGCAACTGCAATGGTTCGAAGGAGAGTTGCAACGTCTTGCAGCACGCGGCTACCGCTGGGAGCACGCCTACACCCAGTGCGTTCTGCAAGCCATGCTCCTCACACTACAACAGAACATAGATCTGAGCAACGTAAACACGAAGATATGTGAACATTTCGCTGATCCAACATTGACTTCTGACACTTCTGAAAGTGAAATTGTTAATCCAGAAATGGAACCAGAAGAGACAGCCTTTATTCGTTATACACCAAACGCTGCCGTCGGGGTTGACTCTGACTTTGCTAATGAAATTTACAACCCACCGATCGTGCCAGAAGATAACCCCAACATAGGTGATATCGAAACGGAAACTCCCAGTGATAAGCTACGAGACTTGATGCTAATGACGGACAATGAAGAGTCTCCTGTTATTTTACCATTTAACGGCTTCAGAGAACGCCTTCAAGCTGAAGGAAAGGGTCGAACCAAAACAAACGTACATCTAAACTTAATCGACAAAGAAAAGAAATCTGTGAATGAAATGATGGCTCAAAACGATCCTAATATCGGATACACAGACGAGGAGCGTTTAGGTGCGCATTTCCATCCGTCCGATATTCCGGAATCTCGGATTTATGACGAAGATTTAGACGAATTGCCTATTGATCCAGACAGCGGGGAGCTAGATGCCTACCGTGCTAAAGGTAAGCCTCCAGGGTTTGAAGGGAAGCATGGCAAAAATATGGATCGTAACATAGTGAATTATATGCGAGATATGATAAAATCTAAACCAAAACAAGATGCCATCTACGCAGAAGGTGGCCCTTTGAATCCTGAAGAATTACAAGCTGAAGATCCAGCTCATACATTAAAAAACAGCAAACATACCGAACATAACCATAATGATTTTGATTATGATCCGGCGTACGCCTACGTTTTACTTAAAAATGGATTACTCACCAATTGGGCTAAAGGAATTTCATTTATATCGAAGATTGAGGAAATGTTGGGTCTGGATAAAAACTCATTCTCGAACCCGCGAGTTGACCTACGGGAAGTGACATTCAAAGTGGAAAAAAATAATAAAGGCTACGATGCTGTTGACGTTGCAAGGCGAGTTGATGACATCAAAGATGATCTATTAAGAGAAACTGGAGCCCAGATAATTTCTACTGGAGTTGGTGACAGAAGCAAGCGTCCTATAATGCGTCATGTAGCTTCTCCGGATACTTTAATTTTCGGCTTAGAGCTACCCGTTTTCCTGGCGTTAGTTGGAAGCTTTTCTGTTTTGGTCGTAGGCGCAATTATTTTCGCTATTCTGTTGAAAAAAGACATGTCCGGAAAGAAAAAATCACAAGGCCTTTCATCTGCAGCGGAAATTGATGCAGAGGCCACAAGAGATTACCAGGAGCTATGTAGAGCACGTATGTCTGGGAAGTGGTCTGGCCAACAGACAGCAGCTAATGCGTCATCTAACCCCGCGGAGCCTGCCCAACGCATCACGTCTTTGTCACGAGAACCCGATGGAAACTCGCCTTCTACACGATCCAGCACCTCTTCATGGAGCGAAGAACCAGCTCTAACCAATATGGACATTTCAACTGGTCACATGGTCTTGGCATATATGGAAGATCACCTTCGCAACAAGGATCGCTTGGAGCAGGAGTGGCGTGCTCTGTGCGCATACGAGGCCGAACCCTGTGCTACAACTTCAGCTCTTAAGCCGGAGAATAGTGCGAAAAACAGATATGCTGATGTGCTTCCTTACGACCATTCCAGGGTTATTCTGAACACCTTATCCAACCATGTCGGATCGGATTACATCAATGCTTCGACAATTACTGACCACGACCCACGCAATCCTGCATACATCGCATCAGCTGGACCACTCCCCCACACGGCACCTGATTTCTGGCAGATGGTCTGGGAGCAAGGCTCAGTTGTCATGGTGATGTTGACTCGACTTACGGAAAACGGACAACAACTGTGCCACCGCTATTGGCCTGAGGAAGGATCTGAACTTTACCACATTTATGAGGTACACCTTGTCAGCGAGCATATTTGGTGTGATGACTACCTGGTTCGAAGCTTCTACCTGAAGAACCAACGTACCGGCGAGACCCGCACCGTCACCCAGTTCCATTTCCTCTCGTGGCCTGAGAACGGAGTACCAGCGTCGACCAAAGCTCTGTTGGAATTCAGGAGAAAGGTGAATAAGTCTTACAGAGGAAGATCTTGTCCAATCGTAGTGCACTGCAGCGATGGCGCCGGCCGCTCTGGAACGTACTGTCTGATAGACATGGTATTGAACCGGATGGCTAAAGGCGCTAAGGAGATCGACATCGCAGCCACTCTCGAGCATATCCGAGACCAGCGCACCCGCACTGTCGCTACTAAGCAGCAATTTGAGTTTGTGCTGATGGCGGTCGCTGAAGAGGATAGACTACTGGGCTTACTAGATAAAAGAAAATACTAA
Protein
MYGLSLRQALWALALINALAPSHADGNIGCLFSAPLCVEGVEWCYDDFAFGKCIPVYEEESEEGALYRYDMGPEQLQWFEGELQRLAARGYRWEHAYTQCVLQAMLLTLQQNIDLSNVNTKICEHFADPTLTSDTSESEIVNPEMEPEETAFIRYTPNAAVGVDSDFANEIYNPPIVPEDNPNIGDIETETPSDKLRDLMLMTDNEESPVILPFNGFRERLQAEGKGRTKTNVHLNLIDKEKKSVNEMMAQNDPNIGYTDEERLGAHFHPSDIPESRIYDEDLDELPIDPDSGELDAYRAKGKPPGFEGKHGKNMDRNIVNYMRDMIKSKPKQDAIYAEGGPLNPEELQAEDPAHTLKNSKHTEHNHNDFDYDPAYAYVLLKNGLLTNWAKGISFISKIEEMLGLDKNSFSNPRVDLREVTFKVEKNNKGYDAVDVARRVDDIKDDLLRETGAQIISTGVGDRSKRPIMRHVASPDTLIFGLELPVFLALVGSFSVLVVGAIIFAILLKKDMSGKKKSQGLSSAAEIDAEATRDYQELCRARMSGKWSGQQTAANASSNPAEPAQRITSLSREPDGNSPSTRSSTSSWSEEPALTNMDISTGHMVLAYMEDHLRNKDRLEQEWRALCAYEAEPCATTSALKPENSAKNRYADVLPYDHSRVILNTLSNHVGSDYINASTITDHDPRNPAYIASAGPLPHTAPDFWQMVWEQGSVVMVMLTRLTENGQQLCHRYWPEEGSELYHIYEVHLVSEHIWCDDYLVRSFYLKNQRTGETRTVTQFHFLSWPENGVPASTKALLEFRRKVNKSYRGRSCPIVVHCSDGAGRSGTYCLIDMVLNRMAKGAKEIDIAATLEHIRDQRTRTVATKQQFEFVLMAVAEEDRLLGLLDKRKY
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
PDB
2QEP
E-value=2.43068e-127,
Score=1170
Ontologies
GO
Topology
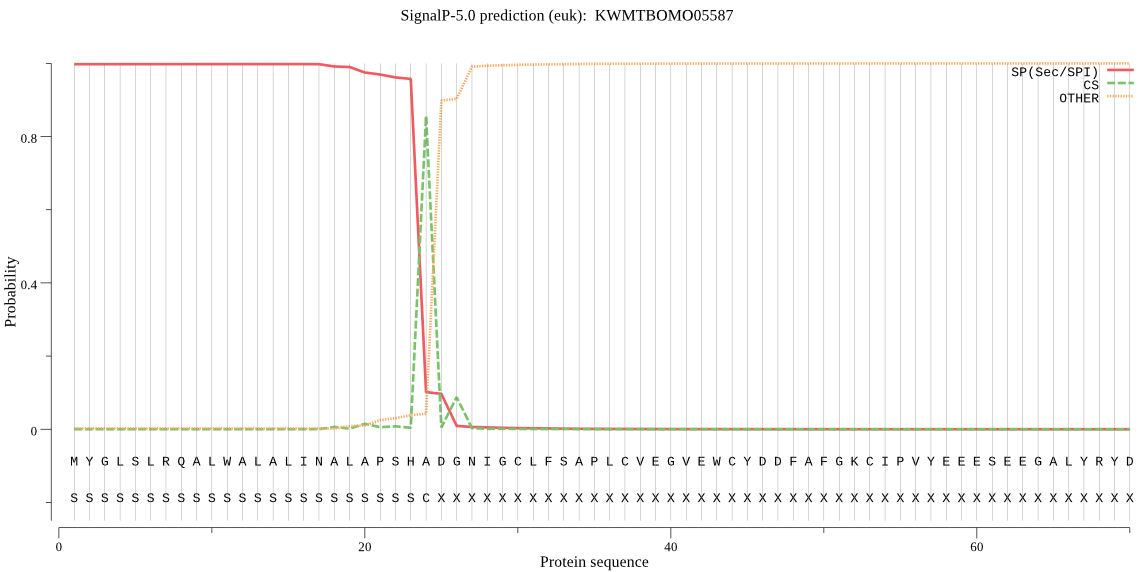
SignalP
Position: 1 - 24,
Likelihood: 0.998000
Length:
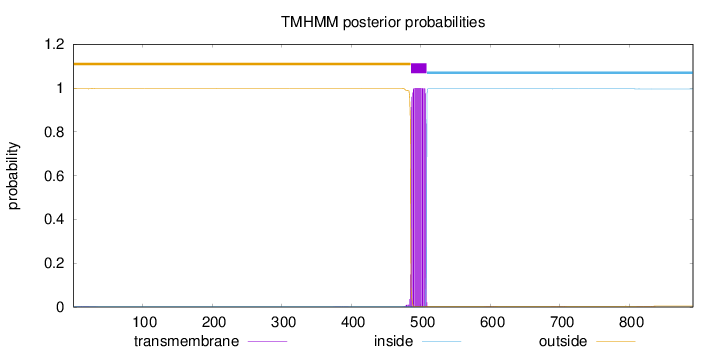
891
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.97118
Exp number, first 60 AAs:
0.03074
Total prob of N-in:
0.00368
outside
1 - 485
TMhelix
486 - 508
inside
509 - 891
Population Genetic Test Statistics
Pi
339.019763
Theta
219.931989
Tajima's D
1.78793
CLR
0.073586
CSRT
0.844607769611519
Interpretation
Uncertain
