Gene
KWMTBOMO05578
Pre Gene Modal
BGIBMGA006586
Annotation
PREDICTED:_alpha-scruin-like_[Plutella_xylostella]
Full name
Beta-scruin
+ More
Alpha-scruin
Influenza virus NS1A-binding protein homolog A
Alpha-scruin
Influenza virus NS1A-binding protein homolog A
Location in the cell
Mitochondrial Reliability : 2.523
Sequence
CDS
ATGAACGTTGCTCGCATGGCGATGGGCTGCTTGAAGTACAGGGACTACATTTGGGTCGCCGGCGGCATGACGGGCGACAAGAAGAGGCCCGTCAGTAAAATCGTGGAGTGCTACAATTCAAGAACCAATGAGTGGACAGAAATACACAATCTCCGTTTTCCTCGATGTTTCACAACTCTGTTTTCAATGAATGACAAACTGTACATTGTCGGCGGAGCAGGGAAAATATCCGAAAAGGAAAAAACACCGAGTAGCGTTGGAGCCATCGATGCTTGGGATTGGAAGGAGCGAGCGTGGAAACTGGAGACGGAGATGTCGATGCCGCGTCACGGACACGCACTCGCCTACCTCGGTACTCAGCTTATAATCATAGGAGGCGTCACTACAATATACATGCGAGCACTGAATAATGTAGAATCTTATTGCTGCGAACGTGGCGCTTGGATCCGTGGGGTGGCAGCCCTACCCTCGCCACTGTCCGGTCACGGGGCAGTTACATTGCCACCTGCTTCGCTTATGTGA
Protein
MNVARMAMGCLKYRDYIWVAGGMTGDKKRPVSKIVECYNSRTNEWTEIHNLRFPRCFTTLFSMNDKLYIVGGAGKISEKEKTPSSVGAIDAWDWKERAWKLETEMSMPRHGHALAYLGTQLIIIGGVTTIYMRALNNVESYCCERGAWIRGVAALPSPLSGHGAVTLPPASLM
Summary
Description
Plays a role in cell division and in the dynamic organization of the actin skeleton as a stabilizer of actin filaments by association with F-actin through Kelch repeats.
Similarity
Belongs to the G-protein coupled receptor 2 family.
Keywords
Kelch repeat
Repeat
Alternative splicing
Complete proteome
Cytoplasm
Cytoskeleton
Nucleus
Reference proteome
Feature
chain Beta-scruin
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JAP1
A0A2A4K267
A0A194PEC4
A0A1C9EGK4
A0A212F1A7
E0VM55
+ More
A0A194QLG1 D6X201 A0A088AQ25 A0A0L7R7F1 A0A154PKK6 A0A310ST73 A0A2A3EIE9 A0A0C9Q771 T1HYJ3 A0A224XJB8 A0A0A9XQR7 A0A0A9XFE2 T1JEX1 A0A1B6D328 A0A1B6D0X8 A0A226F0K4 A0A1B6IAI1 A0A1B6L3Y5 A0A2R7WG18 A0A1B6ELB0 A0A1S3JCD5 A0A1S3JIH9 A0A0M8ZW88 A0A2P8Y214 Q25386 Q25390 A0A1D2MGV4 C3ZAN5 A0A2T7NUD0 H2ZIF7 A0A2T7NSQ7 A0A1A9XE02 A0A1A9ULC2 B7Q735 A0A1B0AN56 A0A1W2WBW9 F6V396 I2EYT4 T1IK99 B7P7S4 B1H341 B7QK30 A0A2D4VKD8 H6CDM2 A0A3B3RVD4 F7E9S0 A0A3B3RV90 A0A1L8GGL0 A0A2C9JHJ4 A0A369AS77 K1PF79 A0A1H3LX36 A0A3B1J6C4 W5LCD6 I3JUM7 A0A2F0BLQ3 A0A1H6LGE8 A0A1A8GGQ9 A0A2D0NIP4 A0A1D2ND91 A0A2P6L5B7 A0A3S0ZGP4 A0A0B6YQQ6 A0A2G9RG49 A0A1B0D3K8 A0A3Q2X2Z9 A0A293L487 A0A2D4JUG9 Q5RG82 A0A3Q1CFF6 A0A3P8ST07 A0A3P8PBG7 A0A3Q4HXC5 V9KFA6 A0A3B4G620 A0A3Q4N4R0 A0A3Q2WR22 A0A3P8PBE6 A0A3P9CQZ9 A0A3Q2WH97 A0A3Q2WGN5 A0A3P9CQZ4 A0A1A8LMJ6
A0A194QLG1 D6X201 A0A088AQ25 A0A0L7R7F1 A0A154PKK6 A0A310ST73 A0A2A3EIE9 A0A0C9Q771 T1HYJ3 A0A224XJB8 A0A0A9XQR7 A0A0A9XFE2 T1JEX1 A0A1B6D328 A0A1B6D0X8 A0A226F0K4 A0A1B6IAI1 A0A1B6L3Y5 A0A2R7WG18 A0A1B6ELB0 A0A1S3JCD5 A0A1S3JIH9 A0A0M8ZW88 A0A2P8Y214 Q25386 Q25390 A0A1D2MGV4 C3ZAN5 A0A2T7NUD0 H2ZIF7 A0A2T7NSQ7 A0A1A9XE02 A0A1A9ULC2 B7Q735 A0A1B0AN56 A0A1W2WBW9 F6V396 I2EYT4 T1IK99 B7P7S4 B1H341 B7QK30 A0A2D4VKD8 H6CDM2 A0A3B3RVD4 F7E9S0 A0A3B3RV90 A0A1L8GGL0 A0A2C9JHJ4 A0A369AS77 K1PF79 A0A1H3LX36 A0A3B1J6C4 W5LCD6 I3JUM7 A0A2F0BLQ3 A0A1H6LGE8 A0A1A8GGQ9 A0A2D0NIP4 A0A1D2ND91 A0A2P6L5B7 A0A3S0ZGP4 A0A0B6YQQ6 A0A2G9RG49 A0A1B0D3K8 A0A3Q2X2Z9 A0A293L487 A0A2D4JUG9 Q5RG82 A0A3Q1CFF6 A0A3P8ST07 A0A3P8PBG7 A0A3Q4HXC5 V9KFA6 A0A3B4G620 A0A3Q4N4R0 A0A3Q2WR22 A0A3P8PBE6 A0A3P9CQZ9 A0A3Q2WH97 A0A3Q2WGN5 A0A3P9CQZ4 A0A1A8LMJ6
Pubmed
EMBL
BABH01008646
BABH01008647
NWSH01000287
PCG77732.1
KQ459606
KPI91617.1
+ More
KX778610 AON96630.1 AGBW02010922 OWR47519.1 DS235289 EEB14461.1 KQ461198 KPJ06209.1 KQ971371 EFA10195.2 KQ414639 KOC66800.1 KQ434936 KZC12004.1 KQ760221 OAD61375.1 KZ288232 PBC31478.1 GBYB01009993 JAG79760.1 ACPB03020118 GFTR01008307 JAW08119.1 GBHO01024154 JAG19450.1 GBHO01024157 GBRD01000588 JAG19447.1 JAG65233.1 JH432130 GEDC01017220 JAS20078.1 GEDC01018015 JAS19283.1 LNIX01000001 OXA62944.1 GECU01023836 JAS83870.1 GEBQ01021525 JAT18452.1 KK854775 PTY18604.1 GECZ01031078 JAS38691.1 KQ435840 KOX71326.1 PYGN01001031 PSN38305.1 Z47541 Z38132 LJIJ01001321 ODM92141.1 GG666602 EEN50438.1 PZQS01000009 PVD24789.1 PVD24186.1 ABJB010120763 ABJB010128823 ABJB010186399 ABJB010399346 ABJB010500663 ABJB010574955 ABJB010662129 ABJB010675563 ABJB010858327 ABJB011008628 DS872016 EEC14657.1 JXJN01000723 JXJN01000724 EAAA01000057 CP002961 AFK04837.1 JH430450 ABJB010017723 ABJB010110186 ABJB010381258 ABJB010457894 ABJB010632335 ABJB010771594 ABJB010918446 ABJB010966307 ABJB010978255 ABJB010990706 DS653188 EEC02646.1 BC161238 AAI61238.1 ABJB010219446 ABJB010643383 ABJB010786116 ABJB010915131 DS956570 EEC19202.1 NYVD01000117 MAB14385.1 JH601044 EHS59321.1 AAMC01009141 AAMC01009142 AAMC01009143 AAMC01009144 AAMC01009145 AAMC01009146 AAMC01009147 AAMC01009148 CM004473 OCT82980.1 QPJT01000023 RCX12202.1 JH816123 EKC20248.1 FNOY01000054 SDY68926.1 AERX01012990 NTJE010292210 MBW03851.1 FNXF01000006 SEH87643.1 HAEB01016195 HAEC01002139 SBQ70216.1 PDUD01000001 PHN08375.1 LJIJ01000088 ODN03055.1 MWRG01001696 PRD33774.1 RQTK01000506 RUS78504.1 HACG01011759 CEK58624.1 KV947546 PIO26241.1 AJVK01002940 GFWV01000737 MAA25467.1 IACL01012384 LAB00098.1 BX957305 BC046068 BC066513 JW863947 AFO96464.1 HAEF01007657 HAEG01005920 SBR45114.1
KX778610 AON96630.1 AGBW02010922 OWR47519.1 DS235289 EEB14461.1 KQ461198 KPJ06209.1 KQ971371 EFA10195.2 KQ414639 KOC66800.1 KQ434936 KZC12004.1 KQ760221 OAD61375.1 KZ288232 PBC31478.1 GBYB01009993 JAG79760.1 ACPB03020118 GFTR01008307 JAW08119.1 GBHO01024154 JAG19450.1 GBHO01024157 GBRD01000588 JAG19447.1 JAG65233.1 JH432130 GEDC01017220 JAS20078.1 GEDC01018015 JAS19283.1 LNIX01000001 OXA62944.1 GECU01023836 JAS83870.1 GEBQ01021525 JAT18452.1 KK854775 PTY18604.1 GECZ01031078 JAS38691.1 KQ435840 KOX71326.1 PYGN01001031 PSN38305.1 Z47541 Z38132 LJIJ01001321 ODM92141.1 GG666602 EEN50438.1 PZQS01000009 PVD24789.1 PVD24186.1 ABJB010120763 ABJB010128823 ABJB010186399 ABJB010399346 ABJB010500663 ABJB010574955 ABJB010662129 ABJB010675563 ABJB010858327 ABJB011008628 DS872016 EEC14657.1 JXJN01000723 JXJN01000724 EAAA01000057 CP002961 AFK04837.1 JH430450 ABJB010017723 ABJB010110186 ABJB010381258 ABJB010457894 ABJB010632335 ABJB010771594 ABJB010918446 ABJB010966307 ABJB010978255 ABJB010990706 DS653188 EEC02646.1 BC161238 AAI61238.1 ABJB010219446 ABJB010643383 ABJB010786116 ABJB010915131 DS956570 EEC19202.1 NYVD01000117 MAB14385.1 JH601044 EHS59321.1 AAMC01009141 AAMC01009142 AAMC01009143 AAMC01009144 AAMC01009145 AAMC01009146 AAMC01009147 AAMC01009148 CM004473 OCT82980.1 QPJT01000023 RCX12202.1 JH816123 EKC20248.1 FNOY01000054 SDY68926.1 AERX01012990 NTJE010292210 MBW03851.1 FNXF01000006 SEH87643.1 HAEB01016195 HAEC01002139 SBQ70216.1 PDUD01000001 PHN08375.1 LJIJ01000088 ODN03055.1 MWRG01001696 PRD33774.1 RQTK01000506 RUS78504.1 HACG01011759 CEK58624.1 KV947546 PIO26241.1 AJVK01002940 GFWV01000737 MAA25467.1 IACL01012384 LAB00098.1 BX957305 BC046068 BC066513 JW863947 AFO96464.1 HAEF01007657 HAEG01005920 SBR45114.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000009046
UP000053240
+ More
UP000007266 UP000005203 UP000053825 UP000076502 UP000242457 UP000015103 UP000198287 UP000085678 UP000053105 UP000245037 UP000094527 UP000001554 UP000245119 UP000007875 UP000092443 UP000078200 UP000001555 UP000092460 UP000008144 UP000002875 UP000005078 UP000261540 UP000008143 UP000186698 UP000076420 UP000253034 UP000005408 UP000198640 UP000018467 UP000005207 UP000199371 UP000223913 UP000271974 UP000092462 UP000264840 UP000000437 UP000257160 UP000265080 UP000265100 UP000261580 UP000261460 UP000265160
UP000007266 UP000005203 UP000053825 UP000076502 UP000242457 UP000015103 UP000198287 UP000085678 UP000053105 UP000245037 UP000094527 UP000001554 UP000245119 UP000007875 UP000092443 UP000078200 UP000001555 UP000092460 UP000008144 UP000002875 UP000005078 UP000261540 UP000008143 UP000186698 UP000076420 UP000253034 UP000005408 UP000198640 UP000018467 UP000005207 UP000199371 UP000223913 UP000271974 UP000092462 UP000264840 UP000000437 UP000257160 UP000265080 UP000265100 UP000261580 UP000261460 UP000265160
Pfam
Interpro
IPR006652
Kelch_1
+ More
IPR015915 Kelch-typ_b-propeller
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR011705 BACK
IPR017096 BTB-kelch_protein
IPR000048 IQ_motif_EF-hand-BS
IPR011498 Kelch_2
IPR029058 AB_hydrolase
IPR038765 Papain-like_cys_pep_sf
IPR002931 Transglutaminase-like
IPR008547 DUF829_TMEM53
IPR030104 IPP
IPR030609 KLHL33
IPR030594 KLHL17
IPR031750 DUF4734
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR011043 Gal_Oxase/kelch_b-propeller
IPR000421 FA58C
IPR013783 Ig-like_fold
IPR008979 Galactose-bd-like_sf
IPR030605 KLHL8
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR000203 GPS
IPR002049 Laminin_EGF
IPR001879 GPCR_2_extracellular_dom
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR032471 GAIN_dom_N
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000832 GPCR_2_secretin-like
IPR017981 GPCR_2-like
IPR001791 Laminin_G
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015915 Kelch-typ_b-propeller
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR011705 BACK
IPR017096 BTB-kelch_protein
IPR000048 IQ_motif_EF-hand-BS
IPR011498 Kelch_2
IPR029058 AB_hydrolase
IPR038765 Papain-like_cys_pep_sf
IPR002931 Transglutaminase-like
IPR008547 DUF829_TMEM53
IPR030104 IPP
IPR030609 KLHL33
IPR030594 KLHL17
IPR031750 DUF4734
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR011043 Gal_Oxase/kelch_b-propeller
IPR000421 FA58C
IPR013783 Ig-like_fold
IPR008979 Galactose-bd-like_sf
IPR030605 KLHL8
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR000203 GPS
IPR002049 Laminin_EGF
IPR001879 GPCR_2_extracellular_dom
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR032471 GAIN_dom_N
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000832 GPCR_2_secretin-like
IPR017981 GPCR_2-like
IPR001791 Laminin_G
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
SUPFAM
CDD
ProteinModelPortal
H9JAP1
A0A2A4K267
A0A194PEC4
A0A1C9EGK4
A0A212F1A7
E0VM55
+ More
A0A194QLG1 D6X201 A0A088AQ25 A0A0L7R7F1 A0A154PKK6 A0A310ST73 A0A2A3EIE9 A0A0C9Q771 T1HYJ3 A0A224XJB8 A0A0A9XQR7 A0A0A9XFE2 T1JEX1 A0A1B6D328 A0A1B6D0X8 A0A226F0K4 A0A1B6IAI1 A0A1B6L3Y5 A0A2R7WG18 A0A1B6ELB0 A0A1S3JCD5 A0A1S3JIH9 A0A0M8ZW88 A0A2P8Y214 Q25386 Q25390 A0A1D2MGV4 C3ZAN5 A0A2T7NUD0 H2ZIF7 A0A2T7NSQ7 A0A1A9XE02 A0A1A9ULC2 B7Q735 A0A1B0AN56 A0A1W2WBW9 F6V396 I2EYT4 T1IK99 B7P7S4 B1H341 B7QK30 A0A2D4VKD8 H6CDM2 A0A3B3RVD4 F7E9S0 A0A3B3RV90 A0A1L8GGL0 A0A2C9JHJ4 A0A369AS77 K1PF79 A0A1H3LX36 A0A3B1J6C4 W5LCD6 I3JUM7 A0A2F0BLQ3 A0A1H6LGE8 A0A1A8GGQ9 A0A2D0NIP4 A0A1D2ND91 A0A2P6L5B7 A0A3S0ZGP4 A0A0B6YQQ6 A0A2G9RG49 A0A1B0D3K8 A0A3Q2X2Z9 A0A293L487 A0A2D4JUG9 Q5RG82 A0A3Q1CFF6 A0A3P8ST07 A0A3P8PBG7 A0A3Q4HXC5 V9KFA6 A0A3B4G620 A0A3Q4N4R0 A0A3Q2WR22 A0A3P8PBE6 A0A3P9CQZ9 A0A3Q2WH97 A0A3Q2WGN5 A0A3P9CQZ4 A0A1A8LMJ6
A0A194QLG1 D6X201 A0A088AQ25 A0A0L7R7F1 A0A154PKK6 A0A310ST73 A0A2A3EIE9 A0A0C9Q771 T1HYJ3 A0A224XJB8 A0A0A9XQR7 A0A0A9XFE2 T1JEX1 A0A1B6D328 A0A1B6D0X8 A0A226F0K4 A0A1B6IAI1 A0A1B6L3Y5 A0A2R7WG18 A0A1B6ELB0 A0A1S3JCD5 A0A1S3JIH9 A0A0M8ZW88 A0A2P8Y214 Q25386 Q25390 A0A1D2MGV4 C3ZAN5 A0A2T7NUD0 H2ZIF7 A0A2T7NSQ7 A0A1A9XE02 A0A1A9ULC2 B7Q735 A0A1B0AN56 A0A1W2WBW9 F6V396 I2EYT4 T1IK99 B7P7S4 B1H341 B7QK30 A0A2D4VKD8 H6CDM2 A0A3B3RVD4 F7E9S0 A0A3B3RV90 A0A1L8GGL0 A0A2C9JHJ4 A0A369AS77 K1PF79 A0A1H3LX36 A0A3B1J6C4 W5LCD6 I3JUM7 A0A2F0BLQ3 A0A1H6LGE8 A0A1A8GGQ9 A0A2D0NIP4 A0A1D2ND91 A0A2P6L5B7 A0A3S0ZGP4 A0A0B6YQQ6 A0A2G9RG49 A0A1B0D3K8 A0A3Q2X2Z9 A0A293L487 A0A2D4JUG9 Q5RG82 A0A3Q1CFF6 A0A3P8ST07 A0A3P8PBG7 A0A3Q4HXC5 V9KFA6 A0A3B4G620 A0A3Q4N4R0 A0A3Q2WR22 A0A3P8PBE6 A0A3P9CQZ9 A0A3Q2WH97 A0A3Q2WGN5 A0A3P9CQZ4 A0A1A8LMJ6
PDB
5YY8
E-value=4.93872e-09,
Score=141
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Associated with actin filaments. With evidence from 1 publications.
Cytoskeleton Associated with actin filaments. With evidence from 1 publications.
Nucleus Associated with actin filaments. With evidence from 1 publications.
Cytoskeleton Associated with actin filaments. With evidence from 1 publications.
Nucleus Associated with actin filaments. With evidence from 1 publications.
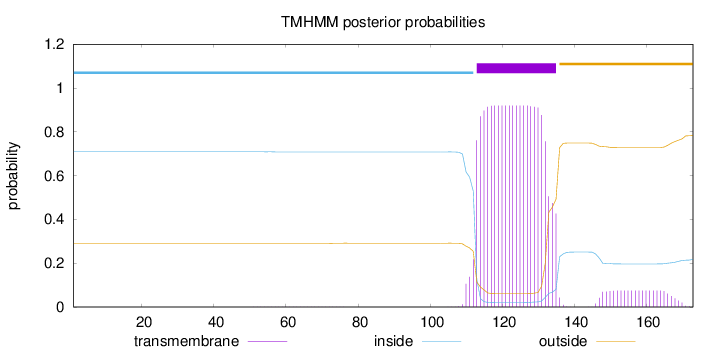
Length:
173
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.49029
Exp number, first 60 AAs:
0.00882
Total prob of N-in:
0.70937
inside
1 - 112
TMhelix
113 - 135
outside
136 - 173
Population Genetic Test Statistics
Pi
186.242997
Theta
146.060268
Tajima's D
0.998148
CLR
0.780022
CSRT
0.654217289135543
Interpretation
Uncertain
