Pre Gene Modal
BGIBMGA006754
Annotation
pelota-like_protein_[Bombyx_mori]
Full name
Protein pelota homolog
+ More
Protein pelota
Protein pelota
Location in the cell
Cytoplasmic Reliability : 2.03 Nuclear Reliability : 1.529
Sequence
CDS
ATGAAACTCGTTCACAAAAACATTGAGAAAGATGACTCCGGGAGTATTGGGCTCATACCGGAGGAACCCGAGGACATGTGGCATGCGTATAATCTAATCACAGAAGGCGATGCTGTAACTGCATCAACTGTTCGGAAAGTTCAAACAGAATCTTCGACCGGGTCTTCCACTAGCAGTAGAGTTAGGACTACGCTCACCATTACTGTAGAAAACATTGACTTTGATACCCAAGCATGTGTGTTAAGGTTGAAAGGTCGTAACATTGTGGAAAATCAATATGTTAAGATGGGTGCATATCACACATTGGATTTAGAACTGAATCGAAAGTTTATTCTGCAGAAGGTATTGTGGGATTCGGTAGCATTAGAACGCGTGGAAACTGCTTGTGATCCAGCAAGTAATGCTGATGTTGCAGCAGTTGTCATGCAGGAAGGACTGGCACATGTCTGTCTTATTACACCCTCAATGACACTTGTGAGGATTAAAATTGACATCACAATACCTAAAAAACGGAAAGGATTTGTACAACAGCATGAAAAGGGCTTAAACAAGTTTTATGAAGCGGTAATGCAAGGAATTCTCAGGCATATAGATTTTAACATTGTCAAGTGTGTAATCATTGCTTCACCTGGTTTTGTGAAGGATCAGTTCTTTGAGTATATGATGCAGCAGGCAATCAAAACAGATAATAAGCTACTTATAGATAACAGAAGTAAATTTTTACTGGTCAAAGCTTCATCTGGATTCAAACATTCTTTAAAAGAAGTTTTACAAGAACCTGCAGTAATGGCAAAAATATCAGATACGAAGGCTGCAAGTGAAGTAAAATTACTGGAAAGTTTTTACACAATGTTGCAGTTAGAGCCCTCAAAAGCGTTCTATGGAAAAAAACATGTAATGAGGGCTAATGAGGCCCTTGCAATTGAAACACTCATGATCTCAGATAAATTGTTCAGGTGTCAAGATACATTGAAGCGGAAAGAGTATGTTGCTCTTGTAGATTCTGTCCGAGAAAATGGTGGAGATGTTAGAATATTCTCTAGTATGCATGTGTCTGGGGAACAGCTAGATCAATTGACGGGTATAGCTGCAGTCCTGCGATTTCCTATGCCGGAACTTGAAGACAGTGATGCTGAAGAAGACAGCGAAAATGATTCTGATTAG
Protein
MKLVHKNIEKDDSGSIGLIPEEPEDMWHAYNLITEGDAVTASTVRKVQTESSTGSSTSSRVRTTLTITVENIDFDTQACVLRLKGRNIVENQYVKMGAYHTLDLELNRKFILQKVLWDSVALERVETACDPASNADVAAVVMQEGLAHVCLITPSMTLVRIKIDITIPKKRKGFVQQHEKGLNKFYEAVMQGILRHIDFNIVKCVIIASPGFVKDQFFEYMMQQAIKTDNKLLIDNRSKFLLVKASSGFKHSLKEVLQEPAVMAKISDTKAASEVKLLESFYTMLQLEPSKAFYGKKHVMRANEALAIETLMISDKLFRCQDTLKRKEYVALVDSVRENGGDVRIFSSMHVSGEQLDQLTGIAAVLRFPMPELEDSDAEEDSENDSD
Summary
Description
May function in recognizing stalled ribosomes and triggering endonucleolytic cleavage of the mRNA, a mechanism to release non-functional ribosomes and degrade damaged mRNAs.
Required prior to the first meiotic division for spindle formation and nuclear envelope breakdown during spermatogenesis. It is also required for normal eye patterning and for mitotic divisions in the ovary. Required for ovarian germ line stem cell self-renewal. May play a role in regulating translation. May function in recognizing stalled ribosomes and triggering endonucleolytic cleavage of the mRNA, a mechanism to release non-functional ribosomes and degrade damaged mRNAs. May have ribonuclease activity (Potential).
Required prior to the first meiotic division for spindle formation and nuclear envelope breakdown during spermatogenesis. It is also required for normal eye patterning and for mitotic divisions in the ovary. Required for ovarian germ line stem cell self-renewal. May play a role in regulating translation. May function in recognizing stalled ribosomes and triggering endonucleolytic cleavage of the mRNA, a mechanism to release non-functional ribosomes and degrade damaged mRNAs. May have ribonuclease activity (Potential).
Cofactor
a divalent metal cation
Similarity
Belongs to the eukaryotic release factor 1 family. Pelota subfamily.
Keywords
Cell cycle
Cell division
Complete proteome
Cytoplasm
Endonuclease
Hydrolase
Meiosis
Metal-binding
Mitosis
Nuclease
Nucleus
Reference proteome
Feature
chain Protein pelota
Uniprot
H9JB59
Q1HQ46
A0A194QRQ0
A0A194PF23
A0A2H1VHC0
A0A2A4K0F9
+ More
A0A1C9EGK1 S4PGA8 A0A0L7L250 V5GPQ1 A0A1W4XI51 A0A1A9XDP3 A0A1A9VZM6 A0A1A9UMU1 A0A1A9ZJ48 D3TRE2 A0A0L0CFT4 A0A1Y1MVS7 A0A182L0Y9 A0A2J7QM97 A0A1S4GYG2 A0A182XL52 A0A182UNT0 A0A182ICH2 A0A1Q3F4Z8 U4TWH5 A0A182QYM9 A0A0K8SXT4 A0A0K8TRP8 A0A1B6KY61 T1PBR8 A0A224XRE3 A0A2M4AJB2 Q170N2 A0A182J8A1 A0A1B6EGQ4 A0A0P4VQJ8 K7IVK8 U5EV94 T1IFI8 B0WGL0 A0A1E1W1H7 A0A2M4CVA9 A0A232FL49 A0A067RQ81 A0A084W4V1 A0A182W6Q4 A0A2M4BR04 B4N1D3 A0A1I8NU07 Q29NB3 B4G7Y7 B3MUX7 B4NY68 E2AA79 A0A2M3ZHJ6 A0A1W4W8S6 B4KIE0 B4LQD1 W8CBU8 B4HW91 B3N8G8 D7ELJ7 A0A2M4CVT5 A0A1B0CSC3 Q7Q477 A0A3B0KIT5 B4Q7V3 A0A0A1XAE6 P48612 E2BMV0 A0A026WKW8 A0A2R5LKR8 A0A1W7R9Z6 A0A2A3EPQ7 A0A131XGK1 A0A088AVL4 A0A0M4E4Y8 A0A224Z559 A0A0K8W0Z8 A0A336LUU1 A0A293MWU4 A0A1L8DS72 G3MNJ2 A0A034WF72 L7MB26 A0A023GJD1 A0A131YNF6 B4JZG6 A0A1E1XFE2 A0A151I6M9 A0A0K8R5W8 A0A151X5M8 A0A158NRH7 A0A195BD03 A0A2I9LPF4 A0A182Y3H0 A0A2M4BQV0 A0A154PPD7 A0A195FSH5
A0A1C9EGK1 S4PGA8 A0A0L7L250 V5GPQ1 A0A1W4XI51 A0A1A9XDP3 A0A1A9VZM6 A0A1A9UMU1 A0A1A9ZJ48 D3TRE2 A0A0L0CFT4 A0A1Y1MVS7 A0A182L0Y9 A0A2J7QM97 A0A1S4GYG2 A0A182XL52 A0A182UNT0 A0A182ICH2 A0A1Q3F4Z8 U4TWH5 A0A182QYM9 A0A0K8SXT4 A0A0K8TRP8 A0A1B6KY61 T1PBR8 A0A224XRE3 A0A2M4AJB2 Q170N2 A0A182J8A1 A0A1B6EGQ4 A0A0P4VQJ8 K7IVK8 U5EV94 T1IFI8 B0WGL0 A0A1E1W1H7 A0A2M4CVA9 A0A232FL49 A0A067RQ81 A0A084W4V1 A0A182W6Q4 A0A2M4BR04 B4N1D3 A0A1I8NU07 Q29NB3 B4G7Y7 B3MUX7 B4NY68 E2AA79 A0A2M3ZHJ6 A0A1W4W8S6 B4KIE0 B4LQD1 W8CBU8 B4HW91 B3N8G8 D7ELJ7 A0A2M4CVT5 A0A1B0CSC3 Q7Q477 A0A3B0KIT5 B4Q7V3 A0A0A1XAE6 P48612 E2BMV0 A0A026WKW8 A0A2R5LKR8 A0A1W7R9Z6 A0A2A3EPQ7 A0A131XGK1 A0A088AVL4 A0A0M4E4Y8 A0A224Z559 A0A0K8W0Z8 A0A336LUU1 A0A293MWU4 A0A1L8DS72 G3MNJ2 A0A034WF72 L7MB26 A0A023GJD1 A0A131YNF6 B4JZG6 A0A1E1XFE2 A0A151I6M9 A0A0K8R5W8 A0A151X5M8 A0A158NRH7 A0A195BD03 A0A2I9LPF4 A0A182Y3H0 A0A2M4BQV0 A0A154PPD7 A0A195FSH5
EC Number
3.1.-.-
Pubmed
19121390
26354079
23622113
26227816
20353571
26108605
+ More
28004739 20966253 12364791 23537049 26369729 25315136 17510324 27129103 20075255 28648823 24845553 24438588 17994087 15632085 17550304 20798317 24495485 18362917 19820115 22936249 25830018 7588080 10731132 12537572 12537569 16280348 24508170 28049606 28797301 22216098 25348373 25576852 26830274 28503490 21347285 29248469 25244985
28004739 20966253 12364791 23537049 26369729 25315136 17510324 27129103 20075255 28648823 24845553 24438588 17994087 15632085 17550304 20798317 24495485 18362917 19820115 22936249 25830018 7588080 10731132 12537572 12537569 16280348 24508170 28049606 28797301 22216098 25348373 25576852 26830274 28503490 21347285 29248469 25244985
EMBL
BABH01008636
DQ443206
ABF51295.1
KQ461198
KPJ06216.1
KQ459606
+ More
KPI91623.1 ODYU01002558 SOQ40233.1 NWSH01000287 PCG77751.1 KX778610 AON96623.1 GAIX01003732 JAA88828.1 JTDY01003554 KOB69349.1 GALX01002367 JAB66099.1 EZ423994 ADD20270.1 JRES01000441 KNC31105.1 GEZM01025376 JAV87487.1 NEVH01013214 PNF29721.1 AAAB01008964 APCN01000799 GFDL01012404 JAV22641.1 KB630423 KB631761 ERL83596.1 ERL85914.1 AXCN02000788 GBRD01007719 JAG58102.1 GDAI01000566 JAI17037.1 GEBQ01023617 GEBQ01018900 GEBQ01010575 GEBQ01007811 JAT16360.1 JAT21077.1 JAT29402.1 JAT32166.1 KA646211 AFP60840.1 GFTR01005713 JAW10713.1 GGFK01007545 MBW40866.1 CH477471 EAT40419.1 GEDC01000199 JAS37099.1 GDKW01001308 JAI55287.1 GANO01001977 JAB57894.1 ACPB03005609 DS231927 EDS27039.1 GDQN01010209 JAT80845.1 GGFL01005017 MBW69195.1 NNAY01000064 OXU31385.1 KK852530 KDR21904.1 ATLV01020383 KE525299 KFB45245.1 GGFJ01006293 MBW55434.1 CH963920 EDW78106.1 CH379060 EAL33429.1 CH479180 EDW28485.1 CH902624 EDV33042.1 CM000157 EDW88670.1 GL438030 EFN69659.1 GGFM01007179 MBW27930.1 CH933807 EDW13437.1 CH940649 EDW63381.1 GAMC01004496 JAC02060.1 CH480818 EDW52286.1 CH954177 EDV58391.1 DS497871 EFA12200.1 GGFL01004770 MBW68948.1 AJWK01025894 AJWK01025895 AJWK01025896 AJWK01025897 AJWK01025898 EAA12313.4 OUUW01000010 SPP85676.1 CM000361 CM002910 EDX04385.1 KMY89301.1 GBXI01006416 JAD07876.1 U27197 AY058644 AE014134 GL449347 EFN82985.1 KK107167 EZA56316.1 GGLE01005781 MBY09907.1 GFAH01000421 JAV47968.1 KZ288198 PBC33697.1 GEFH01001937 JAP66644.1 CP012523 ALC39071.1 GFPF01011035 MAA22181.1 GDHF01007491 JAI44823.1 UFQT01000215 SSX21812.1 GFWV01018907 MAA43635.1 GFDF01004884 JAV09200.1 JO843443 AEO35060.1 GAKP01006177 JAC52775.1 GACK01004685 JAA60349.1 GBBM01001361 JAC34057.1 GEDV01007768 JAP80789.1 CH916379 EDV94088.1 GFAC01001319 JAT97869.1 KQ978465 KYM93754.1 GADI01007550 JAA66258.1 KQ982498 KYQ55687.1 ADTU01024042 KQ976511 KYM82451.1 GFWZ01000280 MBW20270.1 GGFJ01006298 MBW55439.1 KQ434998 KZC13304.1 KQ981280 KYN43391.1
KPI91623.1 ODYU01002558 SOQ40233.1 NWSH01000287 PCG77751.1 KX778610 AON96623.1 GAIX01003732 JAA88828.1 JTDY01003554 KOB69349.1 GALX01002367 JAB66099.1 EZ423994 ADD20270.1 JRES01000441 KNC31105.1 GEZM01025376 JAV87487.1 NEVH01013214 PNF29721.1 AAAB01008964 APCN01000799 GFDL01012404 JAV22641.1 KB630423 KB631761 ERL83596.1 ERL85914.1 AXCN02000788 GBRD01007719 JAG58102.1 GDAI01000566 JAI17037.1 GEBQ01023617 GEBQ01018900 GEBQ01010575 GEBQ01007811 JAT16360.1 JAT21077.1 JAT29402.1 JAT32166.1 KA646211 AFP60840.1 GFTR01005713 JAW10713.1 GGFK01007545 MBW40866.1 CH477471 EAT40419.1 GEDC01000199 JAS37099.1 GDKW01001308 JAI55287.1 GANO01001977 JAB57894.1 ACPB03005609 DS231927 EDS27039.1 GDQN01010209 JAT80845.1 GGFL01005017 MBW69195.1 NNAY01000064 OXU31385.1 KK852530 KDR21904.1 ATLV01020383 KE525299 KFB45245.1 GGFJ01006293 MBW55434.1 CH963920 EDW78106.1 CH379060 EAL33429.1 CH479180 EDW28485.1 CH902624 EDV33042.1 CM000157 EDW88670.1 GL438030 EFN69659.1 GGFM01007179 MBW27930.1 CH933807 EDW13437.1 CH940649 EDW63381.1 GAMC01004496 JAC02060.1 CH480818 EDW52286.1 CH954177 EDV58391.1 DS497871 EFA12200.1 GGFL01004770 MBW68948.1 AJWK01025894 AJWK01025895 AJWK01025896 AJWK01025897 AJWK01025898 EAA12313.4 OUUW01000010 SPP85676.1 CM000361 CM002910 EDX04385.1 KMY89301.1 GBXI01006416 JAD07876.1 U27197 AY058644 AE014134 GL449347 EFN82985.1 KK107167 EZA56316.1 GGLE01005781 MBY09907.1 GFAH01000421 JAV47968.1 KZ288198 PBC33697.1 GEFH01001937 JAP66644.1 CP012523 ALC39071.1 GFPF01011035 MAA22181.1 GDHF01007491 JAI44823.1 UFQT01000215 SSX21812.1 GFWV01018907 MAA43635.1 GFDF01004884 JAV09200.1 JO843443 AEO35060.1 GAKP01006177 JAC52775.1 GACK01004685 JAA60349.1 GBBM01001361 JAC34057.1 GEDV01007768 JAP80789.1 CH916379 EDV94088.1 GFAC01001319 JAT97869.1 KQ978465 KYM93754.1 GADI01007550 JAA66258.1 KQ982498 KYQ55687.1 ADTU01024042 KQ976511 KYM82451.1 GFWZ01000280 MBW20270.1 GGFJ01006298 MBW55439.1 KQ434998 KZC13304.1 KQ981280 KYN43391.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000192223
+ More
UP000092443 UP000091820 UP000078200 UP000092445 UP000037069 UP000075882 UP000235965 UP000076407 UP000075903 UP000075840 UP000030742 UP000075886 UP000095301 UP000008820 UP000075880 UP000002358 UP000015103 UP000002320 UP000215335 UP000027135 UP000030765 UP000075920 UP000007798 UP000095300 UP000001819 UP000008744 UP000007801 UP000002282 UP000000311 UP000192221 UP000009192 UP000008792 UP000001292 UP000008711 UP000007266 UP000092461 UP000007062 UP000268350 UP000000304 UP000000803 UP000008237 UP000053097 UP000242457 UP000005203 UP000092553 UP000001070 UP000078542 UP000075809 UP000005205 UP000078540 UP000076408 UP000076502 UP000078541
UP000092443 UP000091820 UP000078200 UP000092445 UP000037069 UP000075882 UP000235965 UP000076407 UP000075903 UP000075840 UP000030742 UP000075886 UP000095301 UP000008820 UP000075880 UP000002358 UP000015103 UP000002320 UP000215335 UP000027135 UP000030765 UP000075920 UP000007798 UP000095300 UP000001819 UP000008744 UP000007801 UP000002282 UP000000311 UP000192221 UP000009192 UP000008792 UP000001292 UP000008711 UP000007266 UP000092461 UP000007062 UP000268350 UP000000304 UP000000803 UP000008237 UP000053097 UP000242457 UP000005203 UP000092553 UP000001070 UP000078542 UP000075809 UP000005205 UP000078540 UP000076408 UP000076502 UP000078541
Interpro
Gene 3D
ProteinModelPortal
H9JB59
Q1HQ46
A0A194QRQ0
A0A194PF23
A0A2H1VHC0
A0A2A4K0F9
+ More
A0A1C9EGK1 S4PGA8 A0A0L7L250 V5GPQ1 A0A1W4XI51 A0A1A9XDP3 A0A1A9VZM6 A0A1A9UMU1 A0A1A9ZJ48 D3TRE2 A0A0L0CFT4 A0A1Y1MVS7 A0A182L0Y9 A0A2J7QM97 A0A1S4GYG2 A0A182XL52 A0A182UNT0 A0A182ICH2 A0A1Q3F4Z8 U4TWH5 A0A182QYM9 A0A0K8SXT4 A0A0K8TRP8 A0A1B6KY61 T1PBR8 A0A224XRE3 A0A2M4AJB2 Q170N2 A0A182J8A1 A0A1B6EGQ4 A0A0P4VQJ8 K7IVK8 U5EV94 T1IFI8 B0WGL0 A0A1E1W1H7 A0A2M4CVA9 A0A232FL49 A0A067RQ81 A0A084W4V1 A0A182W6Q4 A0A2M4BR04 B4N1D3 A0A1I8NU07 Q29NB3 B4G7Y7 B3MUX7 B4NY68 E2AA79 A0A2M3ZHJ6 A0A1W4W8S6 B4KIE0 B4LQD1 W8CBU8 B4HW91 B3N8G8 D7ELJ7 A0A2M4CVT5 A0A1B0CSC3 Q7Q477 A0A3B0KIT5 B4Q7V3 A0A0A1XAE6 P48612 E2BMV0 A0A026WKW8 A0A2R5LKR8 A0A1W7R9Z6 A0A2A3EPQ7 A0A131XGK1 A0A088AVL4 A0A0M4E4Y8 A0A224Z559 A0A0K8W0Z8 A0A336LUU1 A0A293MWU4 A0A1L8DS72 G3MNJ2 A0A034WF72 L7MB26 A0A023GJD1 A0A131YNF6 B4JZG6 A0A1E1XFE2 A0A151I6M9 A0A0K8R5W8 A0A151X5M8 A0A158NRH7 A0A195BD03 A0A2I9LPF4 A0A182Y3H0 A0A2M4BQV0 A0A154PPD7 A0A195FSH5
A0A1C9EGK1 S4PGA8 A0A0L7L250 V5GPQ1 A0A1W4XI51 A0A1A9XDP3 A0A1A9VZM6 A0A1A9UMU1 A0A1A9ZJ48 D3TRE2 A0A0L0CFT4 A0A1Y1MVS7 A0A182L0Y9 A0A2J7QM97 A0A1S4GYG2 A0A182XL52 A0A182UNT0 A0A182ICH2 A0A1Q3F4Z8 U4TWH5 A0A182QYM9 A0A0K8SXT4 A0A0K8TRP8 A0A1B6KY61 T1PBR8 A0A224XRE3 A0A2M4AJB2 Q170N2 A0A182J8A1 A0A1B6EGQ4 A0A0P4VQJ8 K7IVK8 U5EV94 T1IFI8 B0WGL0 A0A1E1W1H7 A0A2M4CVA9 A0A232FL49 A0A067RQ81 A0A084W4V1 A0A182W6Q4 A0A2M4BR04 B4N1D3 A0A1I8NU07 Q29NB3 B4G7Y7 B3MUX7 B4NY68 E2AA79 A0A2M3ZHJ6 A0A1W4W8S6 B4KIE0 B4LQD1 W8CBU8 B4HW91 B3N8G8 D7ELJ7 A0A2M4CVT5 A0A1B0CSC3 Q7Q477 A0A3B0KIT5 B4Q7V3 A0A0A1XAE6 P48612 E2BMV0 A0A026WKW8 A0A2R5LKR8 A0A1W7R9Z6 A0A2A3EPQ7 A0A131XGK1 A0A088AVL4 A0A0M4E4Y8 A0A224Z559 A0A0K8W0Z8 A0A336LUU1 A0A293MWU4 A0A1L8DS72 G3MNJ2 A0A034WF72 L7MB26 A0A023GJD1 A0A131YNF6 B4JZG6 A0A1E1XFE2 A0A151I6M9 A0A0K8R5W8 A0A151X5M8 A0A158NRH7 A0A195BD03 A0A2I9LPF4 A0A182Y3H0 A0A2M4BQV0 A0A154PPD7 A0A195FSH5
PDB
5LZZ
E-value=2.5446e-140,
Score=1278
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
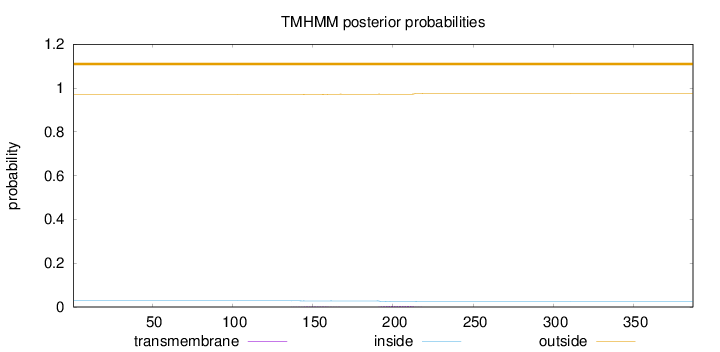
Length:
387
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12553
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02870
outside
1 - 387
Population Genetic Test Statistics
Pi
140.82228
Theta
171.797746
Tajima's D
-0.582069
CLR
12.377029
CSRT
0.217039148042598
Interpretation
Uncertain
