Gene
KWMTBOMO05564 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006749
Annotation
elongation_factor_Tu_[Bombyx_mori]
Full name
Elongation factor Tu
Location in the cell
Cytoplasmic Reliability : 2.25 Mitochondrial Reliability : 2.138
Sequence
CDS
ATGGCTTCAATAACATTTGTGAAAAGTTTGGTTAGTCCAGCTGTGAAATTAGTTCTGAAGCAAAGCCAGTGTGCAAATGTACGAAACACAACAGTCCTGTCAGGATTGACTCCATTAAGTATAATACTCAAGAGACATTATGCAGAAAAACAAATCTTTGAACGTACAAAGCCCCATTGCAATGTTGGCACAATCGGACATGTTGATCACGGGAAGACTACACTAACAGCAGCAATAACTAAGGTATTATCTGATCTCAATCTGGCACAGAAAAAAGGTTATGCAGATATTGACAATGCCCCTGAGGAGAAGGCTCGAGGTATCACTATTAATGTGGCTCATGTCGAATACCAAACCGAACAGAGGCATTATGGACACACTGACTGCCCAGGTCATGCTGACTACATTAAGAACATGATTACAGGCACAGCACAAATGGATGGTGCTATATTAGTAGTAGCTGCAACTGATGGTGTAATGCCACAAACACGCGAACATTTACTACTTGCCAAACAGATTGGTATTCAACATGTGGTGGTGTTCATAAACAAAGTGGATGCCGCTGATGAGGAAATGGTAGAACTGGTTGAGATGGAGATCAGAGAGTTGATGACTGAGATGGGATATGATGGTGACAAGATACCTGTTATTAAAGGTTCTGCTCTCTGTGCTCTTGAAGGCAAAAGTCCTGAAATAGGGGCTGATGCTATCACTAAACTTCTTAAAGAAGTGGACACATTCATTCCAACACCTATCCGAGAGCTCGAAAAACCTTTCCTGATGCCAGTTGAGTCTGTTCACTCAATTCCTGGACGAGGAACTGTCATTACAGGCCGTTTATACAGAGGTGTCTTGAAGAAGGGTACTGATTGTGAAATTGTTGGTCATGGTAAAACAATGAAAACAACCGTAACCGGAGTTGAAATGTTCCACAAAACCTTGGAGGAAGCACAAGCCGGAGATCAACTTGGAGCTCTTGTGCGTTCCATCAAAAGGGAACAAATCAAACGCGGTATGGTCATGGCGAAGCCCGGTACTGCTAAAGCCCATGACAACTTGGAAGCCGCTGTATACATACTTAGCAAGGAGGAAGGCGGTAGATCGAAACCGTTCACATCATACATTCAGTTACAAATGTTTTCAATGACCTGGGACTGCGCCAGTCAAGTTACCATTCCTGAAAAGGAAATGGTAATGCCCGGCGAGGACGCTACGTTAAAACTGAAACTCTTAAAACCAATGGTGTGCGAAACAGGCCAAAGATTCACACTCCGCCTTGGAGACATTACTTTAGGTACTGGAGTCATTACTAAAATAAACAATAATCTATCTGAGGAAGAAAGAAGCAAACTAATGGAAGGCAAGAAGGCGCGAGAAAAGGCAGCCGCAAAGAAGTAA
Protein
MASITFVKSLVSPAVKLVLKQSQCANVRNTTVLSGLTPLSIILKRHYAEKQIFERTKPHCNVGTIGHVDHGKTTLTAAITKVLSDLNLAQKKGYADIDNAPEEKARGITINVAHVEYQTEQRHYGHTDCPGHADYIKNMITGTAQMDGAILVVAATDGVMPQTREHLLLAKQIGIQHVVVFINKVDAADEEMVELVEMEIRELMTEMGYDGDKIPVIKGSALCALEGKSPEIGADAITKLLKEVDTFIPTPIRELEKPFLMPVESVHSIPGRGTVITGRLYRGVLKKGTDCEIVGHGKTMKTTVTGVEMFHKTLEEAQAGDQLGALVRSIKREQIKRGMVMAKPGTAKAHDNLEAAVYILSKEEGGRSKPFTSYIQLQMFSMTWDCASQVTIPEKEMVMPGEDATLKLKLLKPMVCETGQRFTLRLGDITLGTGVITKINNNLSEEERSKLMEGKKAREKAAAKK
Summary
Description
This protein promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis.
Similarity
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EF-Tu/EF-1A subfamily.
Uniprot
Q2F6A6
H9JB54
S4PLV3
A0A1C9EGJ8
A0A3S2ND36
G1C2J8
+ More
A0A212ENC5 A0A2H1X2I4 A0A194QL14 A0A194PKK7 A0A1J1HQB0 J9HFE1 A0A182JTG3 A0A182P133 A0A182GF15 Q16YK5 B0WRV9 A0A182Y315 A0A182WWM2 A0A182V9M5 A0A182TIL4 A0A182I6V8 A0A182J1G3 A0A084WRQ2 Q7QIP5 A0A182MHQ6 A0A2M3Z1K2 A0A2M3Z1V0 A0A2M4A4A0 A0A2M4BMJ5 A0A182FGC1 A0A2M4BMF5 A0A182Q8D0 A0A2M4BM46 W5J9T4 T1DSS7 A0A2M4BMA0 A0A2M4BM45 A0A182R9U8 A0A0L0C7B8 A0A0K8TSH2 A0A023ETK6 A0A0M5J4W0 A0A182WHX7 A0A182NJT7 U5ESU5 A0A1A9ZPI0 A0A1A9WIY1 A0A1B0GBE5 A0A1I8NVN0 A0A3B0J3E9 A0A1L8ECW5 B3NRK1 A0A1A9VJD7 A0A2J7QJE0 A0A0T6B4E2 A0A1L8ED39 B4KMK6 A0A1B0B4H0 B3MGZ5 B4MS85 B4P4G7 B4MCM2 A0A1B0BZW3 B4JYK2 A0A1W4VGG2 A0A0J9RBZ5 Q28ZS9 B4GGU1 A1Z9E3 A0A1A9XHX0 Q86NS6 A0A067R0M6 A0A0K8WEK5 A0A0A1WLQ1 A0A034WVA3 A0A1I8N2B2 D3TM80 D6X0G1 A0A1B0DQV9 V5I8W6 T1P7S4 W8BY98 A0A336LPD7 A0A0P5TUV2 A0A0N7ZLR2 A0A0P4XTN0 A0A0P6G890 A0A0C9QMJ4 A0A0P5FUV1 A0A0P5QIE8 A0A1Y1JRP4 A0A3R7QM95 A0A1L8DRI0 A0A1L8DR91 N6U1S4 J3JU35 A0A1B6E6U5 A0A0P4VIJ7 R4G7T2
A0A212ENC5 A0A2H1X2I4 A0A194QL14 A0A194PKK7 A0A1J1HQB0 J9HFE1 A0A182JTG3 A0A182P133 A0A182GF15 Q16YK5 B0WRV9 A0A182Y315 A0A182WWM2 A0A182V9M5 A0A182TIL4 A0A182I6V8 A0A182J1G3 A0A084WRQ2 Q7QIP5 A0A182MHQ6 A0A2M3Z1K2 A0A2M3Z1V0 A0A2M4A4A0 A0A2M4BMJ5 A0A182FGC1 A0A2M4BMF5 A0A182Q8D0 A0A2M4BM46 W5J9T4 T1DSS7 A0A2M4BMA0 A0A2M4BM45 A0A182R9U8 A0A0L0C7B8 A0A0K8TSH2 A0A023ETK6 A0A0M5J4W0 A0A182WHX7 A0A182NJT7 U5ESU5 A0A1A9ZPI0 A0A1A9WIY1 A0A1B0GBE5 A0A1I8NVN0 A0A3B0J3E9 A0A1L8ECW5 B3NRK1 A0A1A9VJD7 A0A2J7QJE0 A0A0T6B4E2 A0A1L8ED39 B4KMK6 A0A1B0B4H0 B3MGZ5 B4MS85 B4P4G7 B4MCM2 A0A1B0BZW3 B4JYK2 A0A1W4VGG2 A0A0J9RBZ5 Q28ZS9 B4GGU1 A1Z9E3 A0A1A9XHX0 Q86NS6 A0A067R0M6 A0A0K8WEK5 A0A0A1WLQ1 A0A034WVA3 A0A1I8N2B2 D3TM80 D6X0G1 A0A1B0DQV9 V5I8W6 T1P7S4 W8BY98 A0A336LPD7 A0A0P5TUV2 A0A0N7ZLR2 A0A0P4XTN0 A0A0P6G890 A0A0C9QMJ4 A0A0P5FUV1 A0A0P5QIE8 A0A1Y1JRP4 A0A3R7QM95 A0A1L8DRI0 A0A1L8DR91 N6U1S4 J3JU35 A0A1B6E6U5 A0A0P4VIJ7 R4G7T2
Pubmed
19121390
23622113
22118469
26354079
17510324
26483478
+ More
25244985 24438588 12364791 14747013 17210077 20920257 23761445 26108605 26369729 24945155 17994087 18057021 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 25830018 25348373 25315136 20353571 18362917 19820115 24495485 28004739 23537049 22516182 27129103
25244985 24438588 12364791 14747013 17210077 20920257 23761445 26108605 26369729 24945155 17994087 18057021 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 25830018 25348373 25315136 20353571 18362917 19820115 24495485 28004739 23537049 22516182 27129103
EMBL
DQ311166
ABD36111.1
BABH01008626
BABH01008627
BABH01008628
GAIX01003840
+ More
JAA88720.1 KX778610 AON96618.1 RSAL01000092 RVE47987.1 JF728817 JF728818 JF728819 AEJ38219.1 AGBW02013697 OWR42951.1 ODYU01012893 SOQ59426.1 KQ461198 KPJ06222.1 KQ459606 KPI91630.1 CVRI01000017 CRK90248.1 CH477298 EJY57527.1 JXUM01059065 JXUM01059066 KQ562033 KXJ76856.1 CH477515 EAT39702.1 DS232061 EDS33524.1 APCN01003702 ATLV01026176 KE525407 KFB52896.1 AAAB01008807 EAA04167.4 AXCM01000491 GGFM01001635 MBW22386.1 GGFM01001667 MBW22418.1 GGFK01002315 MBW35636.1 GGFJ01005020 MBW54161.1 GGFJ01005033 MBW54174.1 AXCN02000522 GGFJ01005019 MBW54160.1 ADMH02002070 ETN59615.1 GAMD01001344 JAB00247.1 GGFJ01005034 MBW54175.1 GGFJ01005018 MBW54159.1 JRES01000803 KNC28288.1 GDAI01000522 JAI17081.1 GAPW01001397 JAC12201.1 CP012524 ALC41631.1 GANO01003090 JAB56781.1 CCAG010013992 OUUW01000001 SPP73762.1 GFDG01002313 JAV16486.1 CH954179 EDV56153.1 NEVH01013556 PNF28697.1 LJIG01009862 KRT82232.1 GFDG01002306 JAV16493.1 CH933808 EDW10853.2 JXJN01008310 CH902619 EDV37913.1 CH963850 EDW74974.1 CM000158 EDW90606.1 CH940659 EDW71410.1 JXJN01023312 CH916377 EDV90764.1 CM002911 KMY93523.1 CM000071 EAL25534.1 CH479183 EDW35711.1 AE013599 AAF58366.1 BT003749 AAO41413.1 KK852824 KDR15448.1 GDHF01003059 JAI49255.1 GBXI01015004 JAC99287.1 GAKP01000700 JAC58252.1 EZ422532 ADD18808.1 KQ971372 EFA09578.1 AJVK01019411 GALX01003970 JAB64496.1 KA644671 AFP59300.1 GAMC01004972 JAC01584.1 UFQT01000100 SSX19906.1 GDIP01122390 JAL81324.1 GDIP01233041 LRGB01001005 JAI90360.1 KZS14129.1 GDIP01247008 JAI76393.1 GDIQ01047519 JAN47218.1 GBYB01001847 JAG71614.1 GDIQ01255668 JAJ96056.1 GDIQ01120194 JAL31532.1 GEZM01102360 GEZM01102359 JAV51899.1 QCYY01000667 ROT83583.1 GFDF01005124 JAV08960.1 GFDF01005125 JAV08959.1 APGK01043512 KB741014 KB632063 ENN75505.1 ERL88437.1 BT126747 AEE61709.1 GEDC01003663 JAS33635.1 GDKW01002855 JAI53740.1 ACPB03024585 GAHY01002177 JAA75333.1
JAA88720.1 KX778610 AON96618.1 RSAL01000092 RVE47987.1 JF728817 JF728818 JF728819 AEJ38219.1 AGBW02013697 OWR42951.1 ODYU01012893 SOQ59426.1 KQ461198 KPJ06222.1 KQ459606 KPI91630.1 CVRI01000017 CRK90248.1 CH477298 EJY57527.1 JXUM01059065 JXUM01059066 KQ562033 KXJ76856.1 CH477515 EAT39702.1 DS232061 EDS33524.1 APCN01003702 ATLV01026176 KE525407 KFB52896.1 AAAB01008807 EAA04167.4 AXCM01000491 GGFM01001635 MBW22386.1 GGFM01001667 MBW22418.1 GGFK01002315 MBW35636.1 GGFJ01005020 MBW54161.1 GGFJ01005033 MBW54174.1 AXCN02000522 GGFJ01005019 MBW54160.1 ADMH02002070 ETN59615.1 GAMD01001344 JAB00247.1 GGFJ01005034 MBW54175.1 GGFJ01005018 MBW54159.1 JRES01000803 KNC28288.1 GDAI01000522 JAI17081.1 GAPW01001397 JAC12201.1 CP012524 ALC41631.1 GANO01003090 JAB56781.1 CCAG010013992 OUUW01000001 SPP73762.1 GFDG01002313 JAV16486.1 CH954179 EDV56153.1 NEVH01013556 PNF28697.1 LJIG01009862 KRT82232.1 GFDG01002306 JAV16493.1 CH933808 EDW10853.2 JXJN01008310 CH902619 EDV37913.1 CH963850 EDW74974.1 CM000158 EDW90606.1 CH940659 EDW71410.1 JXJN01023312 CH916377 EDV90764.1 CM002911 KMY93523.1 CM000071 EAL25534.1 CH479183 EDW35711.1 AE013599 AAF58366.1 BT003749 AAO41413.1 KK852824 KDR15448.1 GDHF01003059 JAI49255.1 GBXI01015004 JAC99287.1 GAKP01000700 JAC58252.1 EZ422532 ADD18808.1 KQ971372 EFA09578.1 AJVK01019411 GALX01003970 JAB64496.1 KA644671 AFP59300.1 GAMC01004972 JAC01584.1 UFQT01000100 SSX19906.1 GDIP01122390 JAL81324.1 GDIP01233041 LRGB01001005 JAI90360.1 KZS14129.1 GDIP01247008 JAI76393.1 GDIQ01047519 JAN47218.1 GBYB01001847 JAG71614.1 GDIQ01255668 JAJ96056.1 GDIQ01120194 JAL31532.1 GEZM01102360 GEZM01102359 JAV51899.1 QCYY01000667 ROT83583.1 GFDF01005124 JAV08960.1 GFDF01005125 JAV08959.1 APGK01043512 KB741014 KB632063 ENN75505.1 ERL88437.1 BT126747 AEE61709.1 GEDC01003663 JAS33635.1 GDKW01002855 JAI53740.1 ACPB03024585 GAHY01002177 JAA75333.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000183832
+ More
UP000008820 UP000075881 UP000075885 UP000069940 UP000249989 UP000002320 UP000076408 UP000076407 UP000075903 UP000075902 UP000075840 UP000075880 UP000030765 UP000007062 UP000075883 UP000069272 UP000075886 UP000000673 UP000075900 UP000037069 UP000092553 UP000075920 UP000075884 UP000092445 UP000091820 UP000092444 UP000095300 UP000268350 UP000008711 UP000078200 UP000235965 UP000009192 UP000092460 UP000007801 UP000007798 UP000002282 UP000008792 UP000001070 UP000192221 UP000001819 UP000008744 UP000000803 UP000092443 UP000027135 UP000095301 UP000007266 UP000092462 UP000076858 UP000283509 UP000019118 UP000030742 UP000015103
UP000008820 UP000075881 UP000075885 UP000069940 UP000249989 UP000002320 UP000076408 UP000076407 UP000075903 UP000075902 UP000075840 UP000075880 UP000030765 UP000007062 UP000075883 UP000069272 UP000075886 UP000000673 UP000075900 UP000037069 UP000092553 UP000075920 UP000075884 UP000092445 UP000091820 UP000092444 UP000095300 UP000268350 UP000008711 UP000078200 UP000235965 UP000009192 UP000092460 UP000007801 UP000007798 UP000002282 UP000008792 UP000001070 UP000192221 UP000001819 UP000008744 UP000000803 UP000092443 UP000027135 UP000095301 UP000007266 UP000092462 UP000076858 UP000283509 UP000019118 UP000030742 UP000015103
Pfam
Interpro
IPR004161
EFTu-like_2
+ More
IPR009000 Transl_B-barrel_sf
IPR033720 EFTU_2
IPR027417 P-loop_NTPase
IPR041709 EF-Tu_GTP-bd
IPR004541 Transl_elong_EFTu/EF1A_bac/org
IPR031157 G_TR_CS
IPR009001 Transl_elong_EF1A/Init_IF2_C
IPR004160 Transl_elong_EFTu/EF1A_C
IPR000795 TF_GTP-bd_dom
IPR005225 Small_GTP-bd_dom
IPR041470 GCP_N
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR042241 GCP_C_sf
IPR012934 Znf_AD
IPR040457 GCP_C
IPR033467 Tesmin/TSO1-like_CXC
IPR005172 CRC
IPR005026 SAPAP
IPR009000 Transl_B-barrel_sf
IPR033720 EFTU_2
IPR027417 P-loop_NTPase
IPR041709 EF-Tu_GTP-bd
IPR004541 Transl_elong_EFTu/EF1A_bac/org
IPR031157 G_TR_CS
IPR009001 Transl_elong_EF1A/Init_IF2_C
IPR004160 Transl_elong_EFTu/EF1A_C
IPR000795 TF_GTP-bd_dom
IPR005225 Small_GTP-bd_dom
IPR041470 GCP_N
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR042241 GCP_C_sf
IPR012934 Znf_AD
IPR040457 GCP_C
IPR033467 Tesmin/TSO1-like_CXC
IPR005172 CRC
IPR005026 SAPAP
Gene 3D
ProteinModelPortal
Q2F6A6
H9JB54
S4PLV3
A0A1C9EGJ8
A0A3S2ND36
G1C2J8
+ More
A0A212ENC5 A0A2H1X2I4 A0A194QL14 A0A194PKK7 A0A1J1HQB0 J9HFE1 A0A182JTG3 A0A182P133 A0A182GF15 Q16YK5 B0WRV9 A0A182Y315 A0A182WWM2 A0A182V9M5 A0A182TIL4 A0A182I6V8 A0A182J1G3 A0A084WRQ2 Q7QIP5 A0A182MHQ6 A0A2M3Z1K2 A0A2M3Z1V0 A0A2M4A4A0 A0A2M4BMJ5 A0A182FGC1 A0A2M4BMF5 A0A182Q8D0 A0A2M4BM46 W5J9T4 T1DSS7 A0A2M4BMA0 A0A2M4BM45 A0A182R9U8 A0A0L0C7B8 A0A0K8TSH2 A0A023ETK6 A0A0M5J4W0 A0A182WHX7 A0A182NJT7 U5ESU5 A0A1A9ZPI0 A0A1A9WIY1 A0A1B0GBE5 A0A1I8NVN0 A0A3B0J3E9 A0A1L8ECW5 B3NRK1 A0A1A9VJD7 A0A2J7QJE0 A0A0T6B4E2 A0A1L8ED39 B4KMK6 A0A1B0B4H0 B3MGZ5 B4MS85 B4P4G7 B4MCM2 A0A1B0BZW3 B4JYK2 A0A1W4VGG2 A0A0J9RBZ5 Q28ZS9 B4GGU1 A1Z9E3 A0A1A9XHX0 Q86NS6 A0A067R0M6 A0A0K8WEK5 A0A0A1WLQ1 A0A034WVA3 A0A1I8N2B2 D3TM80 D6X0G1 A0A1B0DQV9 V5I8W6 T1P7S4 W8BY98 A0A336LPD7 A0A0P5TUV2 A0A0N7ZLR2 A0A0P4XTN0 A0A0P6G890 A0A0C9QMJ4 A0A0P5FUV1 A0A0P5QIE8 A0A1Y1JRP4 A0A3R7QM95 A0A1L8DRI0 A0A1L8DR91 N6U1S4 J3JU35 A0A1B6E6U5 A0A0P4VIJ7 R4G7T2
A0A212ENC5 A0A2H1X2I4 A0A194QL14 A0A194PKK7 A0A1J1HQB0 J9HFE1 A0A182JTG3 A0A182P133 A0A182GF15 Q16YK5 B0WRV9 A0A182Y315 A0A182WWM2 A0A182V9M5 A0A182TIL4 A0A182I6V8 A0A182J1G3 A0A084WRQ2 Q7QIP5 A0A182MHQ6 A0A2M3Z1K2 A0A2M3Z1V0 A0A2M4A4A0 A0A2M4BMJ5 A0A182FGC1 A0A2M4BMF5 A0A182Q8D0 A0A2M4BM46 W5J9T4 T1DSS7 A0A2M4BMA0 A0A2M4BM45 A0A182R9U8 A0A0L0C7B8 A0A0K8TSH2 A0A023ETK6 A0A0M5J4W0 A0A182WHX7 A0A182NJT7 U5ESU5 A0A1A9ZPI0 A0A1A9WIY1 A0A1B0GBE5 A0A1I8NVN0 A0A3B0J3E9 A0A1L8ECW5 B3NRK1 A0A1A9VJD7 A0A2J7QJE0 A0A0T6B4E2 A0A1L8ED39 B4KMK6 A0A1B0B4H0 B3MGZ5 B4MS85 B4P4G7 B4MCM2 A0A1B0BZW3 B4JYK2 A0A1W4VGG2 A0A0J9RBZ5 Q28ZS9 B4GGU1 A1Z9E3 A0A1A9XHX0 Q86NS6 A0A067R0M6 A0A0K8WEK5 A0A0A1WLQ1 A0A034WVA3 A0A1I8N2B2 D3TM80 D6X0G1 A0A1B0DQV9 V5I8W6 T1P7S4 W8BY98 A0A336LPD7 A0A0P5TUV2 A0A0N7ZLR2 A0A0P4XTN0 A0A0P6G890 A0A0C9QMJ4 A0A0P5FUV1 A0A0P5QIE8 A0A1Y1JRP4 A0A3R7QM95 A0A1L8DRI0 A0A1L8DR91 N6U1S4 J3JU35 A0A1B6E6U5 A0A0P4VIJ7 R4G7T2
PDB
1XB2
E-value=5.90688e-152,
Score=1379
Ontologies
GO
Topology
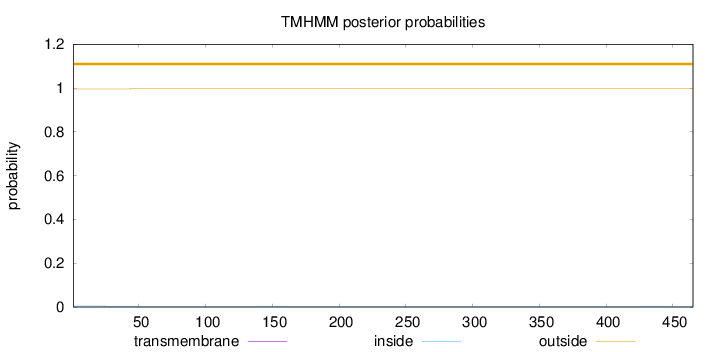
Length:
465
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02654
Exp number, first 60 AAs:
0.02091
Total prob of N-in:
0.00431
outside
1 - 465
Population Genetic Test Statistics
Pi
196.604741
Theta
196.483136
Tajima's D
0.142378
CLR
0.377035
CSRT
0.407529623518824
Interpretation
Uncertain
