Gene
KWMTBOMO05561
Pre Gene Modal
BGIBMGA006592
Annotation
PREDICTED:_kelch_domain-containing_protein_10_homolog_[Papilio_polytes]
Location in the cell
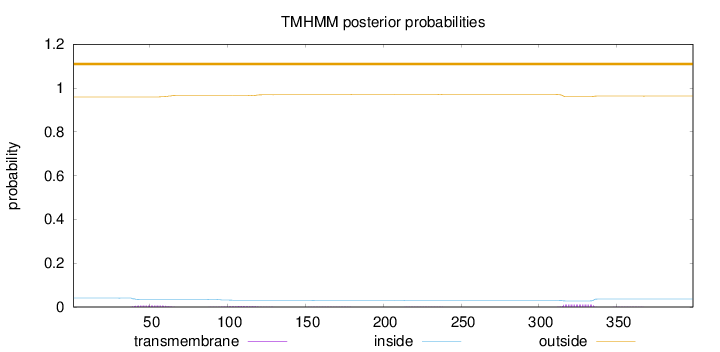
Extracellular Reliability : 1.104
Sequence
CDS
ATGACATCACAGCGAAAAGATTATGTTTTCGAACCTTTTAAGGTAACCGAAGTGAAGTGTCGAGGTTTTGATTTTCCTAGGCCTAGAAGTGGACACAGAATTGCCTGCGACGATGTGAATTTATACTGTTTCGGAGGCTATAATCCGTTTTTACCTTTAGCAAACATAGACCTTGTGAATCCTACTTGGACATCAGCAAGACCTTTGTTTAAAGAGCTTTGGAGTTTTTCTATTGCCACTCGAAAGTGGAAGTTACACAAGGGATATGTAAACATGCCCGACGAATTGGCTTCTAATGCTATGTGTATTAATGGGCGGTATTTAATGATTTTTGGTGGCACAGGTGCACCATTTGGCAACAGATGTAGTAATGATGTCATTGTATGGAGGACTGGGTCAAAAGAAGCTAAGTTACAAATTCTAGATGTAACAGGAAACAGACCGCCTGGACAGTATGGCCAATCTGTTTTATGCCATGATGGTGGATTTTATGCTATAGGAGGCACCAACGGTTTTGCATATAACTGTGATATTTTCAGATTAGATTTAAGGACATTAGTTTGGGAACCAATATTTGTAGGAACTGGCCAAGAAGGTGAACCATTAGGACGATATAGACATGAAATAGCTCAAGTTGGTGATAATTTCTATGTCATTGGAGGAGGAACAGGAGATTTGGCTTTTGAATTAATGGAAATACCAGTTTTTAATCTCACTACAAAATCATGGACAATATTATCGGCCAAACCTGATGATAGTATGACAAATACAATAGCACCTTTACCGAGAAAATGTCATAGTGCTGTTCAAATAGATTCACCAAATGGTGTTCAAGTGTTTGTAGCAGGTGGTTCAGATGGTCAAGCGGTTTTTGAAGATATCTGGAGGTTAGACATTGCTGATCTGCAATGGACCCTTATGAAAAAAACTCAGTTACCTCATCCTTTGTACTTCCATTCATCAACAGTCACGTCCTCTGGTTGCATGTATATATTTGGTGGGATTGAGCCTAAAGAAGATGCAAACTGTAGAAATAATATTTTGTATAAAGTATGGTTATGTATACCAAAACTCAGTGAAATTTGTTGGGAAGCACTAACTTCATTCCATCCTAATCTAGACCAAGTTGACAGAAATACATTACTAAACACTGGTGTACCATTACACTTTGTAAATAGATTACATCCAGACTGGAAATAA
Protein
MTSQRKDYVFEPFKVTEVKCRGFDFPRPRSGHRIACDDVNLYCFGGYNPFLPLANIDLVNPTWTSARPLFKELWSFSIATRKWKLHKGYVNMPDELASNAMCINGRYLMIFGGTGAPFGNRCSNDVIVWRTGSKEAKLQILDVTGNRPPGQYGQSVLCHDGGFYAIGGTNGFAYNCDIFRLDLRTLVWEPIFVGTGQEGEPLGRYRHEIAQVGDNFYVIGGGTGDLAFELMEIPVFNLTTKSWTILSAKPDDSMTNTIAPLPRKCHSAVQIDSPNGVQVFVAGGSDGQAVFEDIWRLDIADLQWTLMKKTQLPHPLYFHSSTVTSSGCMYIFGGIEPKEDANCRNNILYKVWLCIPKLSEICWEALTSFHPNLDQVDRNTLLNTGVPLHFVNRLHPDWK
Summary
Uniprot
A0A2A4J907
A0A2H1X1C8
A0A1C9EGJ9
A0A1E1VXG3
H9JAP7
A0A194PF33
+ More
A0A194QMN7 A0A0L7LB83 A0A212EN87 A0A1E1WRY4 A0A026WLI4 A0A1Y1LJN0 A0A1W4WJ89 A0A2A3EK61 A0A088AVP0 A0A0N1ITS3 V9IH67 D6X316 A0A1Y1LPV2 A0A1W4W7L6 A0A1Q3F518 A0A158NF38 B0VYZ7 A0A1W7R798 E9IFL8 A0A0L7RIY8 A0A023ES92 A0A1S4FV12 Q16NJ1 A0A195C620 F4WEK0 A0A0T6AX15 A0A232F6I4 E2B3I5 K7IVI7 A0A2P8XFU4 A0A154PCV0 A0A067REF1 A0A0M3QVH1 A0A182N1U7 A0A182SM32 Q7Q7V5 A0A2J7R7R7 A0A182JPG6 A0A182HQQ5 A0A195AUW0 A0A182XJG2 A0A2C9H5U7 J3JX04 A0A084WDV1 A0A1L8DBR3 A0A182PZR3 A0A1L8DNR1 A0A182PBB7 A0A182WM53 U5ETG6 A0A182IL44 B4LNJ3 A0A2M4ALN9 A0A2M4AL64 A0A2M4CX12 A0A2M4AM39 W5JCG1 A0A2M4BNN3 A0A3F2YQA1 E2AE06 A0A1B6MI30 A0A182REQ5 A0A182YLH5 A0A182TEK3 B4J5T8 A0A1W4VQ95 A0A1L8E9Z3 B4KM94 A0A1L8EHV0 B4GIY5 Q28YR9 A0A1L8E803 B3MDG8 A0A336L8J8 A0A3B0J2W3 A0A0A1WDL9 B4NMY5 A0A1B0D4S3 A0A195E815 A0A0L0C9K2 T1PA17 U4U7K0 N6UKE4 A0A0C9QXL6 A0A0Q5VUX4 A0A1I8MY50 A0A1I8MY41 Q95TX5 A0A0K8WIT4 B3NMT2 A0A1D2NEA0 A0A034VJD1
A0A194QMN7 A0A0L7LB83 A0A212EN87 A0A1E1WRY4 A0A026WLI4 A0A1Y1LJN0 A0A1W4WJ89 A0A2A3EK61 A0A088AVP0 A0A0N1ITS3 V9IH67 D6X316 A0A1Y1LPV2 A0A1W4W7L6 A0A1Q3F518 A0A158NF38 B0VYZ7 A0A1W7R798 E9IFL8 A0A0L7RIY8 A0A023ES92 A0A1S4FV12 Q16NJ1 A0A195C620 F4WEK0 A0A0T6AX15 A0A232F6I4 E2B3I5 K7IVI7 A0A2P8XFU4 A0A154PCV0 A0A067REF1 A0A0M3QVH1 A0A182N1U7 A0A182SM32 Q7Q7V5 A0A2J7R7R7 A0A182JPG6 A0A182HQQ5 A0A195AUW0 A0A182XJG2 A0A2C9H5U7 J3JX04 A0A084WDV1 A0A1L8DBR3 A0A182PZR3 A0A1L8DNR1 A0A182PBB7 A0A182WM53 U5ETG6 A0A182IL44 B4LNJ3 A0A2M4ALN9 A0A2M4AL64 A0A2M4CX12 A0A2M4AM39 W5JCG1 A0A2M4BNN3 A0A3F2YQA1 E2AE06 A0A1B6MI30 A0A182REQ5 A0A182YLH5 A0A182TEK3 B4J5T8 A0A1W4VQ95 A0A1L8E9Z3 B4KM94 A0A1L8EHV0 B4GIY5 Q28YR9 A0A1L8E803 B3MDG8 A0A336L8J8 A0A3B0J2W3 A0A0A1WDL9 B4NMY5 A0A1B0D4S3 A0A195E815 A0A0L0C9K2 T1PA17 U4U7K0 N6UKE4 A0A0C9QXL6 A0A0Q5VUX4 A0A1I8MY50 A0A1I8MY41 Q95TX5 A0A0K8WIT4 B3NMT2 A0A1D2NEA0 A0A034VJD1
Pubmed
19121390
26354079
26227816
22118469
24508170
30249741
+ More
28004739 18362917 19820115 21347285 21282665 24945155 17510324 21719571 28648823 20798317 20075255 29403074 24845553 12364791 14747013 17210077 22516182 24438588 17994087 20920257 23761445 25244985 15632085 25830018 26108605 23537049 25315136 27289101 25348373
28004739 18362917 19820115 21347285 21282665 24945155 17510324 21719571 28648823 20798317 20075255 29403074 24845553 12364791 14747013 17210077 22516182 24438588 17994087 20920257 23761445 25244985 15632085 25830018 26108605 23537049 25315136 27289101 25348373
EMBL
NWSH01002311
PCG68555.1
ODYU01012704
SOQ59145.1
KX778610
AON96615.1
+ More
GDQN01011621 JAT79433.1 BABH01008626 KQ459606 KPI91633.1 KQ461198 KPJ06225.1 JTDY01001877 KOB72655.1 AGBW02013697 OWR42954.1 GDQN01001483 JAT89571.1 KK107154 QOIP01000001 EZA56905.1 RLU27514.1 GEZM01055577 JAV73018.1 KZ288220 PBC32193.1 KQ435745 KOX76601.1 JR044094 AEY59841.1 KQ971372 EFA10306.1 GEZM01055578 JAV73017.1 GFDL01012384 JAV22661.1 ADTU01013728 DS231813 EDS25738.1 GEHC01000624 JAV47021.1 GL762865 EFZ20633.1 KQ414582 KOC70825.1 GAPW01001478 JAC12120.1 CH477822 EAT35926.1 KQ978287 KYM95621.1 GL888103 EGI67363.1 LJIG01022666 KRT79358.1 NNAY01000837 OXU26255.1 GL445346 EFN89737.1 PYGN01002302 PSN30842.1 KQ434874 KZC09725.1 KK852556 KDR21423.1 CP012524 ALC42409.1 AAAB01008948 EAA10479.4 NEVH01006727 PNF36871.1 APCN01004678 KQ976736 KYM76028.1 BT127772 AEE62734.1 ATLV01023073 KE525340 KFB48395.1 GFDF01010183 JAV03901.1 AXCN02000971 GFDF01005998 JAV08086.1 GANO01002788 JAB57083.1 CH940648 EDW62173.1 GGFK01008370 MBW41691.1 GGFK01008151 MBW41472.1 GGFL01005694 MBW69872.1 GGFK01008519 MBW41840.1 ADMH02001573 ETN62027.1 GGFJ01005410 MBW54551.1 GL438827 EFN68239.1 GEBQ01004380 JAT35597.1 CH916367 EDW01864.1 GFDG01003416 JAV15383.1 CH933808 EDW08758.1 GFDG01000479 JAV18320.1 CH479183 EDW36403.1 CM000071 EAL25897.2 GFDG01004087 JAV14712.1 CH902619 EDV36416.2 UFQS01002298 UFQT01002298 SSX13724.1 SSX33145.1 OUUW01000001 SPP75854.1 GBXI01017198 JAC97093.1 CH964282 EDW85724.1 AJVK01024819 AJVK01024820 KQ979479 KYN21355.1 JRES01000729 KNC28931.1 KA645528 KA645529 AFP60158.1 KB632136 ERL89042.1 APGK01029469 KB740735 ENN79147.1 GBYB01005417 JAG75184.1 CH954179 KQS62089.1 AY058460 AAL13689.1 GDHF01001340 JAI50974.1 EDV55086.1 LJIJ01000074 ODN03425.1 GAKP01015546 JAC43406.1
GDQN01011621 JAT79433.1 BABH01008626 KQ459606 KPI91633.1 KQ461198 KPJ06225.1 JTDY01001877 KOB72655.1 AGBW02013697 OWR42954.1 GDQN01001483 JAT89571.1 KK107154 QOIP01000001 EZA56905.1 RLU27514.1 GEZM01055577 JAV73018.1 KZ288220 PBC32193.1 KQ435745 KOX76601.1 JR044094 AEY59841.1 KQ971372 EFA10306.1 GEZM01055578 JAV73017.1 GFDL01012384 JAV22661.1 ADTU01013728 DS231813 EDS25738.1 GEHC01000624 JAV47021.1 GL762865 EFZ20633.1 KQ414582 KOC70825.1 GAPW01001478 JAC12120.1 CH477822 EAT35926.1 KQ978287 KYM95621.1 GL888103 EGI67363.1 LJIG01022666 KRT79358.1 NNAY01000837 OXU26255.1 GL445346 EFN89737.1 PYGN01002302 PSN30842.1 KQ434874 KZC09725.1 KK852556 KDR21423.1 CP012524 ALC42409.1 AAAB01008948 EAA10479.4 NEVH01006727 PNF36871.1 APCN01004678 KQ976736 KYM76028.1 BT127772 AEE62734.1 ATLV01023073 KE525340 KFB48395.1 GFDF01010183 JAV03901.1 AXCN02000971 GFDF01005998 JAV08086.1 GANO01002788 JAB57083.1 CH940648 EDW62173.1 GGFK01008370 MBW41691.1 GGFK01008151 MBW41472.1 GGFL01005694 MBW69872.1 GGFK01008519 MBW41840.1 ADMH02001573 ETN62027.1 GGFJ01005410 MBW54551.1 GL438827 EFN68239.1 GEBQ01004380 JAT35597.1 CH916367 EDW01864.1 GFDG01003416 JAV15383.1 CH933808 EDW08758.1 GFDG01000479 JAV18320.1 CH479183 EDW36403.1 CM000071 EAL25897.2 GFDG01004087 JAV14712.1 CH902619 EDV36416.2 UFQS01002298 UFQT01002298 SSX13724.1 SSX33145.1 OUUW01000001 SPP75854.1 GBXI01017198 JAC97093.1 CH964282 EDW85724.1 AJVK01024819 AJVK01024820 KQ979479 KYN21355.1 JRES01000729 KNC28931.1 KA645528 KA645529 AFP60158.1 KB632136 ERL89042.1 APGK01029469 KB740735 ENN79147.1 GBYB01005417 JAG75184.1 CH954179 KQS62089.1 AY058460 AAL13689.1 GDHF01001340 JAI50974.1 EDV55086.1 LJIJ01000074 ODN03425.1 GAKP01015546 JAC43406.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000053097 UP000279307 UP000192223 UP000242457 UP000005203 UP000053105 UP000007266 UP000005205 UP000002320 UP000053825 UP000008820 UP000078542 UP000007755 UP000215335 UP000008237 UP000002358 UP000245037 UP000076502 UP000027135 UP000092553 UP000075884 UP000075901 UP000007062 UP000235965 UP000075881 UP000075840 UP000078540 UP000076407 UP000075903 UP000030765 UP000075886 UP000075885 UP000075920 UP000075880 UP000008792 UP000000673 UP000069272 UP000000311 UP000075900 UP000076408 UP000075902 UP000001070 UP000192221 UP000009192 UP000008744 UP000001819 UP000007801 UP000268350 UP000007798 UP000092462 UP000078492 UP000037069 UP000030742 UP000019118 UP000008711 UP000095301 UP000094527
UP000053097 UP000279307 UP000192223 UP000242457 UP000005203 UP000053105 UP000007266 UP000005205 UP000002320 UP000053825 UP000008820 UP000078542 UP000007755 UP000215335 UP000008237 UP000002358 UP000245037 UP000076502 UP000027135 UP000092553 UP000075884 UP000075901 UP000007062 UP000235965 UP000075881 UP000075840 UP000078540 UP000076407 UP000075903 UP000030765 UP000075886 UP000075885 UP000075920 UP000075880 UP000008792 UP000000673 UP000069272 UP000000311 UP000075900 UP000076408 UP000075902 UP000001070 UP000192221 UP000009192 UP000008744 UP000001819 UP000007801 UP000268350 UP000007798 UP000092462 UP000078492 UP000037069 UP000030742 UP000019118 UP000008711 UP000095301 UP000094527
Pfam
Interpro
IPR015915
Kelch-typ_b-propeller
+ More
IPR006652 Kelch_1
IPR011043 Gal_Oxase/kelch_b-propeller
IPR011498 Kelch_2
IPR038727 NadR/Ttd14_AAA_dom
IPR033469 CYTH-like_dom_sf
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR009010 Asp_de-COase-like_dom_sf
IPR000734 TAG_lipase
IPR039812 Vesicle-fus_ATPase
IPR029067 CDC48_domain_2-like_sf
IPR003959 ATPase_AAA_core
IPR004201 Cdc48_dom2
IPR013818 Lipase/vitellogenin
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003338 CDC4_N-term_subdom
IPR029058 AB_hydrolase
IPR006652 Kelch_1
IPR011043 Gal_Oxase/kelch_b-propeller
IPR011498 Kelch_2
IPR038727 NadR/Ttd14_AAA_dom
IPR033469 CYTH-like_dom_sf
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR009010 Asp_de-COase-like_dom_sf
IPR000734 TAG_lipase
IPR039812 Vesicle-fus_ATPase
IPR029067 CDC48_domain_2-like_sf
IPR003959 ATPase_AAA_core
IPR004201 Cdc48_dom2
IPR013818 Lipase/vitellogenin
IPR003960 ATPase_AAA_CS
IPR027417 P-loop_NTPase
IPR003338 CDC4_N-term_subdom
IPR029058 AB_hydrolase
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4J907
A0A2H1X1C8
A0A1C9EGJ9
A0A1E1VXG3
H9JAP7
A0A194PF33
+ More
A0A194QMN7 A0A0L7LB83 A0A212EN87 A0A1E1WRY4 A0A026WLI4 A0A1Y1LJN0 A0A1W4WJ89 A0A2A3EK61 A0A088AVP0 A0A0N1ITS3 V9IH67 D6X316 A0A1Y1LPV2 A0A1W4W7L6 A0A1Q3F518 A0A158NF38 B0VYZ7 A0A1W7R798 E9IFL8 A0A0L7RIY8 A0A023ES92 A0A1S4FV12 Q16NJ1 A0A195C620 F4WEK0 A0A0T6AX15 A0A232F6I4 E2B3I5 K7IVI7 A0A2P8XFU4 A0A154PCV0 A0A067REF1 A0A0M3QVH1 A0A182N1U7 A0A182SM32 Q7Q7V5 A0A2J7R7R7 A0A182JPG6 A0A182HQQ5 A0A195AUW0 A0A182XJG2 A0A2C9H5U7 J3JX04 A0A084WDV1 A0A1L8DBR3 A0A182PZR3 A0A1L8DNR1 A0A182PBB7 A0A182WM53 U5ETG6 A0A182IL44 B4LNJ3 A0A2M4ALN9 A0A2M4AL64 A0A2M4CX12 A0A2M4AM39 W5JCG1 A0A2M4BNN3 A0A3F2YQA1 E2AE06 A0A1B6MI30 A0A182REQ5 A0A182YLH5 A0A182TEK3 B4J5T8 A0A1W4VQ95 A0A1L8E9Z3 B4KM94 A0A1L8EHV0 B4GIY5 Q28YR9 A0A1L8E803 B3MDG8 A0A336L8J8 A0A3B0J2W3 A0A0A1WDL9 B4NMY5 A0A1B0D4S3 A0A195E815 A0A0L0C9K2 T1PA17 U4U7K0 N6UKE4 A0A0C9QXL6 A0A0Q5VUX4 A0A1I8MY50 A0A1I8MY41 Q95TX5 A0A0K8WIT4 B3NMT2 A0A1D2NEA0 A0A034VJD1
A0A194QMN7 A0A0L7LB83 A0A212EN87 A0A1E1WRY4 A0A026WLI4 A0A1Y1LJN0 A0A1W4WJ89 A0A2A3EK61 A0A088AVP0 A0A0N1ITS3 V9IH67 D6X316 A0A1Y1LPV2 A0A1W4W7L6 A0A1Q3F518 A0A158NF38 B0VYZ7 A0A1W7R798 E9IFL8 A0A0L7RIY8 A0A023ES92 A0A1S4FV12 Q16NJ1 A0A195C620 F4WEK0 A0A0T6AX15 A0A232F6I4 E2B3I5 K7IVI7 A0A2P8XFU4 A0A154PCV0 A0A067REF1 A0A0M3QVH1 A0A182N1U7 A0A182SM32 Q7Q7V5 A0A2J7R7R7 A0A182JPG6 A0A182HQQ5 A0A195AUW0 A0A182XJG2 A0A2C9H5U7 J3JX04 A0A084WDV1 A0A1L8DBR3 A0A182PZR3 A0A1L8DNR1 A0A182PBB7 A0A182WM53 U5ETG6 A0A182IL44 B4LNJ3 A0A2M4ALN9 A0A2M4AL64 A0A2M4CX12 A0A2M4AM39 W5JCG1 A0A2M4BNN3 A0A3F2YQA1 E2AE06 A0A1B6MI30 A0A182REQ5 A0A182YLH5 A0A182TEK3 B4J5T8 A0A1W4VQ95 A0A1L8E9Z3 B4KM94 A0A1L8EHV0 B4GIY5 Q28YR9 A0A1L8E803 B3MDG8 A0A336L8J8 A0A3B0J2W3 A0A0A1WDL9 B4NMY5 A0A1B0D4S3 A0A195E815 A0A0L0C9K2 T1PA17 U4U7K0 N6UKE4 A0A0C9QXL6 A0A0Q5VUX4 A0A1I8MY50 A0A1I8MY41 Q95TX5 A0A0K8WIT4 B3NMT2 A0A1D2NEA0 A0A034VJD1
PDB
6DO5
E-value=3.31035e-12,
Score=173
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48726
Exp number, first 60 AAs:
0.13506
Total prob of N-in:
0.04078
outside
1 - 399
Population Genetic Test Statistics
Pi
239.755234
Theta
197.805698
Tajima's D
-0.43406
CLR
2.764609
CSRT
0.257937103144843
Interpretation
Uncertain
