Gene
KWMTBOMO05556
Pre Gene Modal
BGIBMGA006599
Annotation
PREDICTED:_laccase-2-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.951
Sequence
CDS
ATGGCGACGAACCTCCTTTTTGAAGACACCACCAAGAATACGGATCGATCGTGTCACCGACCTTGCAAGTTCAACGTGAAACCGAGACGATGTCAGTACAAAATTAATATCGAACCATCCATAATGGAAGATGGAACTCCTGCACTAACCATGAATGGATTAATCCCGGGACCTCCAATTGTCGCTTGCGTCAATGATATAATAATAGTGGAAGTGACCAACAGAATCCCGGGCCAGGATCTAGCGATTCATTGGGCTTCGTCTCCTGGTACTTTCTTCTATCATGCAGACTCAGTTACTCATCAAAGCGATGGACTGTACGGGTCTCTGATAGTCAATCAACCTCAACCTCTGGAGGCTCATTCGAATTTGTATGATTATGACAGGAGCGAAGAAACAACGGTTCTTATTGCCGCCAAATTCGATCACTTCATTACCGGGAATCTCGAAGACTTAAGTCGTTTGAAACCGTATTCTCTAATTGTGAATGGCAATGAAGAAAGTTCTAAATTCTTCGTACTACCAGACTACGCGTACAGATTGAGGCTTATTAACGCAATTGCTATTGAATGTCCCATTGTAATGAGCATTGATAAACACGATGTTGTGATCATTGCCACCGACGGAGAACCAGTCAAGCCTGTATCCACGAGAAAGGTTCAACTGTATCCGGGTGAACGCATGGATGTCGTGGTTAGAGCAGACCAGCCAAGCAACGGATACTGGATAAGAATCAGCGGACAAGGAGTCTGTGGCGGACTCAGTAGCAAATCTATGCTCATATACTCTGGATTCAACTATACTTCTATGTTACAAAATCAGGATGAAAAATTGCAAGACACCGAATTGGATGAAAATACGCCAGCGTTAAATGGACAAGCACTGGAATCGATTCGCGACGATTCTTTCAGTTCCGATATTAAATCGATCTATTTATCGATTGACAGAAAGAAGCCTGTTAAAGAGGAAAATGATTTCAGATACATCAGTGATGCTATTCCAAAGAAGGCTTACTATCCAGCTCCACTATTTCTTCAAGACAAAGGTGTAGTACAAATAAATGAGAAGAGTTTTCTTTACCCGAACGCTCCAATTTTACTGAAGCCTCGGGAAATTAAAGACAATTTGATTTGTAACGTCGGGGATGAGAGCAAGCAATTGGAAATACAATGTATACAGACGGTGGAAGCTGTCGTTGGAGATATGTTGGAGATAGCTTTAATTAATGAAGGAAATGGAAGTGATGAATCCTATACTTTCCACATGCACGGTTTTGGCATGAAGGTGCTAGCAACGGGACATAGCCAAGATGGAAATCCCTTGACGAAAGAGAAGTTTATTCAATTGGACAAGGAGGGGAAACTAATGAGGAATCTACAGAGCCCTCCTATCAAAGACACGATATCCGTTCCTAATAAGGGTTTCGTAATAGTCAGGATCAAACTAAACCGCGGAGGTAGTTGGTTACTCGAATGTAGAGCTTGTTCTGTTTCGTCTCTACCCACAGCAGTCCTGATAAACGTTCCACAGGCATTACCCACATCCATTGTGGATTCATTACCAAAGTGTGGCAGCTATAGACCTCCCGATGTTTTACTTAATTAG
Protein
MATNLLFEDTTKNTDRSCHRPCKFNVKPRRCQYKINIEPSIMEDGTPALTMNGLIPGPPIVACVNDIIIVEVTNRIPGQDLAIHWASSPGTFFYHADSVTHQSDGLYGSLIVNQPQPLEAHSNLYDYDRSEETTVLIAAKFDHFITGNLEDLSRLKPYSLIVNGNEESSKFFVLPDYAYRLRLINAIAIECPIVMSIDKHDVVIIATDGEPVKPVSTRKVQLYPGERMDVVVRADQPSNGYWIRISGQGVCGGLSSKSMLIYSGFNYTSMLQNQDEKLQDTELDENTPALNGQALESIRDDSFSSDIKSIYLSIDRKKPVKEENDFRYISDAIPKKAYYPAPLFLQDKGVVQINEKSFLYPNAPILLKPREIKDNLICNVGDESKQLEIQCIQTVEAVVGDMLEIALINEGNGSDESYTFHMHGFGMKVLATGHSQDGNPLTKEKFIQLDKEGKLMRNLQSPPIKDTISVPNKGFVIVRIKLNRGGSWLLECRACSVSSLPTAVLINVPQALPTSIVDSLPKCGSYRPPDVLLN
Summary
Similarity
Belongs to the multicopper oxidase family.
Uniprot
H9JAQ4
A0A2H1VVE9
A0A2A4J2P4
A0A194PEE7
A0A291ID65
A0A212F4U4
+ More
A0A0L7LUQ7 A0A2J7QYW3 A0A067QZF8 A0A026WHJ0 A0A3L8DYZ1 T1I108 A0A0C9Q220 D6WE75 Q49I41 M4GPQ6 A0A210Q3M7 A0A0J9R7F0 D5MRE2 A1Z6F4 N6U9Q6 A0A212ET43 Q86P50 A0A194PXL3 A0A2W1BMF7 A0A2A4K7B1 A0A0L7RDJ8 A0A1W4XIF1 A0A0N8BE45 A0A0P5SEH3 A0A0P5VN40 A0A0F7LJ96 E1ZWT8 A0A1S3JY25 A0A0P5SWM3 A0A0P6CZG5 A0A0P5TNK5 A0A0P5LTX1 A0A0N8CHB3 A0A0P5TC11 A0A0P5TVL7 A0A0L7R085 A0A154PKC4 A0A0P6AWC6 T1PG09 A0A0C9R4T4 A0A0P6JN49 A0A1I8MAX1 A0A0P5Y202 A0A0P4XSC8 A0A0P5E4P7 A0A0P5YWD4 A0A1S3HLI0 A0A0P5DD91 A0A0P5D266 A0A1I8QC41 A0A0P5CTT9 A0A0N7ZV53 A0A164YF06 A0A0P5XEI2 A0A0N8CHJ7 A0A0P4XGH4 A0A087ZPF2 A0A0P4Y7V8 A0A0P5ZF44 A0A0P5T8S1 A0A0P5ZP30 A0A0P5D482 A0A0P4XLY4 A0A0P5XM91 A0A0P6ES77 A0A0P5EMF1 A0A0N8DDD2 A0A0P5DE71 A0A0P4ZCE1 A0A0P4Y4Z7 A0A0P5E6U6 A0A0N8A3J4 A0A0P5EHJ0 A0A0P4Y7D9 A0A0P5CDY8 A0A067R8J6 A0A0P5D8G5 A0A0P5TKY7 A0A0N7ZU22 A0A0P5BSJ0 A0A0P5TY61 A0A0P5BGH1 A0A0P5T506 A0A0P5CTM8 A0A0P5TYW3 A0A0N8AHB9 A0A0L7RGD6 A0A0N7ZPG5 A0A0P5E6R2 A0A3S2N8B0
A0A0L7LUQ7 A0A2J7QYW3 A0A067QZF8 A0A026WHJ0 A0A3L8DYZ1 T1I108 A0A0C9Q220 D6WE75 Q49I41 M4GPQ6 A0A210Q3M7 A0A0J9R7F0 D5MRE2 A1Z6F4 N6U9Q6 A0A212ET43 Q86P50 A0A194PXL3 A0A2W1BMF7 A0A2A4K7B1 A0A0L7RDJ8 A0A1W4XIF1 A0A0N8BE45 A0A0P5SEH3 A0A0P5VN40 A0A0F7LJ96 E1ZWT8 A0A1S3JY25 A0A0P5SWM3 A0A0P6CZG5 A0A0P5TNK5 A0A0P5LTX1 A0A0N8CHB3 A0A0P5TC11 A0A0P5TVL7 A0A0L7R085 A0A154PKC4 A0A0P6AWC6 T1PG09 A0A0C9R4T4 A0A0P6JN49 A0A1I8MAX1 A0A0P5Y202 A0A0P4XSC8 A0A0P5E4P7 A0A0P5YWD4 A0A1S3HLI0 A0A0P5DD91 A0A0P5D266 A0A1I8QC41 A0A0P5CTT9 A0A0N7ZV53 A0A164YF06 A0A0P5XEI2 A0A0N8CHJ7 A0A0P4XGH4 A0A087ZPF2 A0A0P4Y7V8 A0A0P5ZF44 A0A0P5T8S1 A0A0P5ZP30 A0A0P5D482 A0A0P4XLY4 A0A0P5XM91 A0A0P6ES77 A0A0P5EMF1 A0A0N8DDD2 A0A0P5DE71 A0A0P4ZCE1 A0A0P4Y4Z7 A0A0P5E6U6 A0A0N8A3J4 A0A0P5EHJ0 A0A0P4Y7D9 A0A0P5CDY8 A0A067R8J6 A0A0P5D8G5 A0A0P5TKY7 A0A0N7ZU22 A0A0P5BSJ0 A0A0P5TY61 A0A0P5BGH1 A0A0P5T506 A0A0P5CTM8 A0A0P5TYW3 A0A0N8AHB9 A0A0L7RGD6 A0A0N7ZPG5 A0A0P5E6R2 A0A3S2N8B0
Pubmed
EMBL
BABH01008620
ODYU01004678
SOQ44815.1
NWSH01003453
PCG66271.1
KQ459606
+ More
KPI91642.1 KY049902 ATG88100.1 AGBW02010309 OWR48754.1 JTDY01000047 KOB79192.1 NEVH01009082 PNF33770.1 KK853076 KDR11753.1 KK107207 EZA55448.1 QOIP01000002 RLU25335.1 ACPB03020385 GBYB01008053 JAG77820.1 KQ971324 EFA01166.1 AY884061 AAX84202.2 JQ292847 AFD33366.1 NEDP02005120 OWF43346.1 CM002911 KMY91619.1 AB514130 BAJ06132.1 AE013599 AAN16124.2 APGK01043587 KB741015 ENN75342.1 AGBW02012655 OWR44647.1 BT003483 AAO39486.1 KQ459588 KPI97763.1 KZ150026 PZC74805.1 NWSH01000053 PCG80165.1 KQ414613 KOC68923.1 GDIQ01186699 JAK65026.1 GDIP01140785 JAL62929.1 GDIP01097790 JAM05925.1 KP318028 AKH61981.1 GL434905 EFN74335.1 GDIP01241174 GDIP01232183 GDIP01230923 GDIP01222079 GDIP01220931 GDIP01185570 GDIP01134237 GDIP01124992 GDIP01064046 LRGB01000915 JAL69477.1 KZS15189.1 GDIQ01087496 JAN07241.1 GDIP01141440 JAL62274.1 GDIQ01165221 JAK86504.1 GDIP01241175 GDIP01232184 GDIP01230924 GDIP01222080 GDIP01220930 GDIP01167326 GDIP01165924 GDIP01147145 GDIP01138874 GDIP01134235 GDIP01131743 GDIP01120831 GDIP01074054 GDIP01064047 GDIP01044646 JAL71971.1 GDIP01210175 GDIP01175564 GDIP01147144 GDIP01146474 GDIP01134236 GDIP01131744 GDIP01130384 GDIP01051957 JAL69478.1 GDIP01126090 JAL77624.1 KQ414672 KOC64253.1 KQ434943 KZC12283.1 GDIP01025604 JAM78111.1 KA647757 AFP62386.1 GBYB01007762 JAG77529.1 GDIQ01013899 JAN80838.1 GDIP01065683 GDIP01065681 GDIP01064253 GDIP01064251 JAM38032.1 GDIP01238723 JAI84678.1 GDIP01165125 JAJ58277.1 GDIP01065682 GDIP01064252 JAM38033.1 GDIP01161677 JAJ61725.1 GDIP01166486 JAJ56916.1 GDIP01166832 JAJ56570.1 GDIP01209513 GDIP01073337 JAJ13889.1 KZS15188.1 GDIP01209514 GDIP01073338 JAM30377.1 GDIP01210710 GDIP01131071 JAL72643.1 GDIP01241462 GDIP01241461 JAI81940.1 GDIP01231267 JAI92134.1 GDIP01239974 GDIP01044794 GDIP01044792 JAM58921.1 GDIP01131070 JAL72644.1 GDIP01041092 JAM62623.1 GDIP01221212 GDIP01161178 JAJ62224.1 GDIP01239973 GDIP01239972 GDIP01044793 JAI83429.1 GDIP01070045 JAM33670.1 GDIQ01071949 JAN22788.1 GDIP01145930 JAJ77472.1 GDIP01044791 JAM58924.1 GDIP01158787 JAJ64615.1 GDIP01227818 JAI95583.1 GDIP01232520 GDIP01232519 JAI90881.1 GDIP01145931 JAJ77471.1 GDIP01185985 JAJ37417.1 GDIP01143028 JAJ80374.1 GDIP01232518 JAI90883.1 GDIP01176778 JAJ46624.1 KK852865 KDR14757.1 GDIP01159679 JAJ63723.1 GDIP01124893 JAL78821.1 GDIP01212561 JAJ10841.1 GDIP01185019 JAJ38383.1 GDIP01124892 JAL78822.1 GDIP01185018 JAJ38384.1 GDIP01137482 JAL66232.1 GDIP01171365 JAJ52037.1 GDIP01137481 JAL66233.1 GDIP01147385 JAJ76017.1 KQ414597 KOC69894.1 GDIP01225417 JAI97984.1 GDIP01147386 JAJ76016.1 RSAL01000212 RVE44181.1
KPI91642.1 KY049902 ATG88100.1 AGBW02010309 OWR48754.1 JTDY01000047 KOB79192.1 NEVH01009082 PNF33770.1 KK853076 KDR11753.1 KK107207 EZA55448.1 QOIP01000002 RLU25335.1 ACPB03020385 GBYB01008053 JAG77820.1 KQ971324 EFA01166.1 AY884061 AAX84202.2 JQ292847 AFD33366.1 NEDP02005120 OWF43346.1 CM002911 KMY91619.1 AB514130 BAJ06132.1 AE013599 AAN16124.2 APGK01043587 KB741015 ENN75342.1 AGBW02012655 OWR44647.1 BT003483 AAO39486.1 KQ459588 KPI97763.1 KZ150026 PZC74805.1 NWSH01000053 PCG80165.1 KQ414613 KOC68923.1 GDIQ01186699 JAK65026.1 GDIP01140785 JAL62929.1 GDIP01097790 JAM05925.1 KP318028 AKH61981.1 GL434905 EFN74335.1 GDIP01241174 GDIP01232183 GDIP01230923 GDIP01222079 GDIP01220931 GDIP01185570 GDIP01134237 GDIP01124992 GDIP01064046 LRGB01000915 JAL69477.1 KZS15189.1 GDIQ01087496 JAN07241.1 GDIP01141440 JAL62274.1 GDIQ01165221 JAK86504.1 GDIP01241175 GDIP01232184 GDIP01230924 GDIP01222080 GDIP01220930 GDIP01167326 GDIP01165924 GDIP01147145 GDIP01138874 GDIP01134235 GDIP01131743 GDIP01120831 GDIP01074054 GDIP01064047 GDIP01044646 JAL71971.1 GDIP01210175 GDIP01175564 GDIP01147144 GDIP01146474 GDIP01134236 GDIP01131744 GDIP01130384 GDIP01051957 JAL69478.1 GDIP01126090 JAL77624.1 KQ414672 KOC64253.1 KQ434943 KZC12283.1 GDIP01025604 JAM78111.1 KA647757 AFP62386.1 GBYB01007762 JAG77529.1 GDIQ01013899 JAN80838.1 GDIP01065683 GDIP01065681 GDIP01064253 GDIP01064251 JAM38032.1 GDIP01238723 JAI84678.1 GDIP01165125 JAJ58277.1 GDIP01065682 GDIP01064252 JAM38033.1 GDIP01161677 JAJ61725.1 GDIP01166486 JAJ56916.1 GDIP01166832 JAJ56570.1 GDIP01209513 GDIP01073337 JAJ13889.1 KZS15188.1 GDIP01209514 GDIP01073338 JAM30377.1 GDIP01210710 GDIP01131071 JAL72643.1 GDIP01241462 GDIP01241461 JAI81940.1 GDIP01231267 JAI92134.1 GDIP01239974 GDIP01044794 GDIP01044792 JAM58921.1 GDIP01131070 JAL72644.1 GDIP01041092 JAM62623.1 GDIP01221212 GDIP01161178 JAJ62224.1 GDIP01239973 GDIP01239972 GDIP01044793 JAI83429.1 GDIP01070045 JAM33670.1 GDIQ01071949 JAN22788.1 GDIP01145930 JAJ77472.1 GDIP01044791 JAM58924.1 GDIP01158787 JAJ64615.1 GDIP01227818 JAI95583.1 GDIP01232520 GDIP01232519 JAI90881.1 GDIP01145931 JAJ77471.1 GDIP01185985 JAJ37417.1 GDIP01143028 JAJ80374.1 GDIP01232518 JAI90883.1 GDIP01176778 JAJ46624.1 KK852865 KDR14757.1 GDIP01159679 JAJ63723.1 GDIP01124893 JAL78821.1 GDIP01212561 JAJ10841.1 GDIP01185019 JAJ38383.1 GDIP01124892 JAL78822.1 GDIP01185018 JAJ38384.1 GDIP01137482 JAL66232.1 GDIP01171365 JAJ52037.1 GDIP01137481 JAL66233.1 GDIP01147385 JAJ76017.1 KQ414597 KOC69894.1 GDIP01225417 JAI97984.1 GDIP01147386 JAJ76016.1 RSAL01000212 RVE44181.1
Proteomes
Interpro
SUPFAM
SSF49503
SSF49503
Gene 3D
ProteinModelPortal
H9JAQ4
A0A2H1VVE9
A0A2A4J2P4
A0A194PEE7
A0A291ID65
A0A212F4U4
+ More
A0A0L7LUQ7 A0A2J7QYW3 A0A067QZF8 A0A026WHJ0 A0A3L8DYZ1 T1I108 A0A0C9Q220 D6WE75 Q49I41 M4GPQ6 A0A210Q3M7 A0A0J9R7F0 D5MRE2 A1Z6F4 N6U9Q6 A0A212ET43 Q86P50 A0A194PXL3 A0A2W1BMF7 A0A2A4K7B1 A0A0L7RDJ8 A0A1W4XIF1 A0A0N8BE45 A0A0P5SEH3 A0A0P5VN40 A0A0F7LJ96 E1ZWT8 A0A1S3JY25 A0A0P5SWM3 A0A0P6CZG5 A0A0P5TNK5 A0A0P5LTX1 A0A0N8CHB3 A0A0P5TC11 A0A0P5TVL7 A0A0L7R085 A0A154PKC4 A0A0P6AWC6 T1PG09 A0A0C9R4T4 A0A0P6JN49 A0A1I8MAX1 A0A0P5Y202 A0A0P4XSC8 A0A0P5E4P7 A0A0P5YWD4 A0A1S3HLI0 A0A0P5DD91 A0A0P5D266 A0A1I8QC41 A0A0P5CTT9 A0A0N7ZV53 A0A164YF06 A0A0P5XEI2 A0A0N8CHJ7 A0A0P4XGH4 A0A087ZPF2 A0A0P4Y7V8 A0A0P5ZF44 A0A0P5T8S1 A0A0P5ZP30 A0A0P5D482 A0A0P4XLY4 A0A0P5XM91 A0A0P6ES77 A0A0P5EMF1 A0A0N8DDD2 A0A0P5DE71 A0A0P4ZCE1 A0A0P4Y4Z7 A0A0P5E6U6 A0A0N8A3J4 A0A0P5EHJ0 A0A0P4Y7D9 A0A0P5CDY8 A0A067R8J6 A0A0P5D8G5 A0A0P5TKY7 A0A0N7ZU22 A0A0P5BSJ0 A0A0P5TY61 A0A0P5BGH1 A0A0P5T506 A0A0P5CTM8 A0A0P5TYW3 A0A0N8AHB9 A0A0L7RGD6 A0A0N7ZPG5 A0A0P5E6R2 A0A3S2N8B0
A0A0L7LUQ7 A0A2J7QYW3 A0A067QZF8 A0A026WHJ0 A0A3L8DYZ1 T1I108 A0A0C9Q220 D6WE75 Q49I41 M4GPQ6 A0A210Q3M7 A0A0J9R7F0 D5MRE2 A1Z6F4 N6U9Q6 A0A212ET43 Q86P50 A0A194PXL3 A0A2W1BMF7 A0A2A4K7B1 A0A0L7RDJ8 A0A1W4XIF1 A0A0N8BE45 A0A0P5SEH3 A0A0P5VN40 A0A0F7LJ96 E1ZWT8 A0A1S3JY25 A0A0P5SWM3 A0A0P6CZG5 A0A0P5TNK5 A0A0P5LTX1 A0A0N8CHB3 A0A0P5TC11 A0A0P5TVL7 A0A0L7R085 A0A154PKC4 A0A0P6AWC6 T1PG09 A0A0C9R4T4 A0A0P6JN49 A0A1I8MAX1 A0A0P5Y202 A0A0P4XSC8 A0A0P5E4P7 A0A0P5YWD4 A0A1S3HLI0 A0A0P5DD91 A0A0P5D266 A0A1I8QC41 A0A0P5CTT9 A0A0N7ZV53 A0A164YF06 A0A0P5XEI2 A0A0N8CHJ7 A0A0P4XGH4 A0A087ZPF2 A0A0P4Y7V8 A0A0P5ZF44 A0A0P5T8S1 A0A0P5ZP30 A0A0P5D482 A0A0P4XLY4 A0A0P5XM91 A0A0P6ES77 A0A0P5EMF1 A0A0N8DDD2 A0A0P5DE71 A0A0P4ZCE1 A0A0P4Y4Z7 A0A0P5E6U6 A0A0N8A3J4 A0A0P5EHJ0 A0A0P4Y7D9 A0A0P5CDY8 A0A067R8J6 A0A0P5D8G5 A0A0P5TKY7 A0A0N7ZU22 A0A0P5BSJ0 A0A0P5TY61 A0A0P5BGH1 A0A0P5T506 A0A0P5CTM8 A0A0P5TYW3 A0A0N8AHB9 A0A0L7RGD6 A0A0N7ZPG5 A0A0P5E6R2 A0A3S2N8B0
PDB
1ASQ
E-value=9.10052e-21,
Score=248
Ontologies
GO
Topology
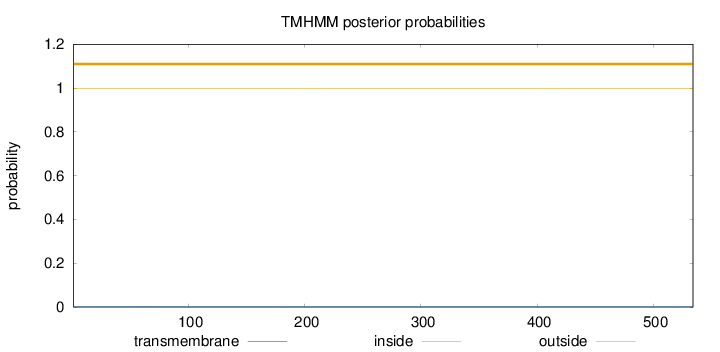
Length:
534
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01133
Exp number, first 60 AAs:
0.00338
Total prob of N-in:
0.00106
outside
1 - 534
Population Genetic Test Statistics
Pi
247.136115
Theta
178.389732
Tajima's D
1.664539
CLR
0.172212
CSRT
0.819859007049648
Interpretation
Uncertain
