Gene
KWMTBOMO05555
Pre Gene Modal
BGIBMGA006747
Annotation
hypothetical_protein_KGM_02850_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 1.375 PlasmaMembrane Reliability : 1.417
Sequence
CDS
ATGGGTGGAATAAATGCAAAACCTCATGCACATCCGTACATCGTATCGTTGCAGATTAGATTTCTGTTTTTAAAATTGCATATTTGTGCAGGCACATTGCTTAACGAATATTGGATCCTGTCTGCCGCCCACTGTATTCCAAGTTTCTTTCTGATCAAATGGCTCCCGATGGAAGCCATTGCAGGATCAAATAATATCAATCATTTGGCACGAACAGCACAGATTGCCACAATTTCAAAAAGAATACCGCATCCGCTATTTACCAGTGACATAGCACCTTATGATATAGCGATGCTTAAAACAAGCAAGCCTTTCACTTTCACAAAAGAAGTTAAACCCATAAATTTACCGGAAAACTCTCAACTTCGACACAAAGACCTTATTCTTGCCGGTTGGGGTGAACTCAGTGAGACGAGCATCATATTTTCTAACTCACCAAGTAAATTGCAGGAAGTGCGAGTGCAATACATCCCGTATAAAGAATGTCACGAAGCTATGGAAAACCTTTTTGAAGACGATGTTAGTATTGAAGACATTGAAACTATGAAAGAAAACGTTCACATTTGTACGGGTCCATTAACGGGTGGAGTGGCAGCCTGCAGTGGAGATTCCGGCGGTCCACTTATTGAATACATATCTCATTATAACAGCGATAATGCAAAAAAGGAATTCAATTATGGTAATGACGTAAATAGTAAAGAAAATTCACGTAGAAGCCAAGATGGAAACGCCGATCATAAAACACCATATATAATCGGAATAGTTTCATGGGGATTCGTACCTTGTGGAGAACCAGGTGCACCCACAGTATTCACGAATGTTTCTCATTACATAGATTTCATTTATGATCATATCTACGAATGA
Protein
MGGINAKPHAHPYIVSLQIRFLFLKLHICAGTLLNEYWILSAAHCIPSFFLIKWLPMEAIAGSNNINHLARTAQIATISKRIPHPLFTSDIAPYDIAMLKTSKPFTFTKEVKPINLPENSQLRHKDLILAGWGELSETSIIFSNSPSKLQEVRVQYIPYKECHEAMENLFEDDVSIEDIETMKENVHICTGPLTGGVAACSGDSGGPLIEYISHYNSDNAKKEFNYGNDVNSKENSRRSQDGNADHKTPYIIGIVSWGFVPCGEPGAPTVFTNVSHYIDFIYDHIYE
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A212F4T3
A0A2A4J497
A0A2H1VVN5
A0A194PF43
A0A194QMP7
A0A0L7LUS5
+ More
W5JNQ1 A0A067RAJ4 E2AY08 E2C9W6 K7IV00 E2C9W4 A0A182TI71 A0A034WBG7 A0A0K8UWD9 E2C9W5 Q7PW17 A0A0K8VEN9 A0A0A1X0J3 A0A0K8U084 A0A195F7K8 A0A0C9S2B8 A0A0L0CPW6 A0A182VL28 A0A182R9M5 T1DPE8 A0A182QML4 A0A182TBT1 A0A182PPS1 A0A232EW13 A0A0C9RXJ7 E0VDU6 A0A084VG87 E5LCR7 B0W1J3 F4X0M1 W8BVD4 A0A182MMN1 D0R8R5 D2A5Z6 A0A232ET09 A0A158NDD6 A0A026WJK5 A0A151WYC0 A0A232EVX7 A0A0C9RRV3 K7IV02 A0A182J6W6 A0A0L7R675 D0R8R1 K7IZJ3 K7IV01 J9EB53 Q16H18 K7J1V4 E9IJV0 Q8T4T5 F4X0M2 B4NG84 A0A182X994 A0A084W0D6 A0A182PJ42 Q9BIG0 A0A1I8MJT5 Q16NR3 A0A0C9R9U5 B4GF17 A0A182QM63 A0A0L0CMG5 A0A1S4GBU1 A0A1I8P599 K7IV03 Q8N0R8 A0A3B0JP11 A0A182IMH4 A0A3L8DI96 A0A1L8DQU0 A0A182L8F6 A0A182GUH7 A0A1W4U884 Q7PX30 Q16NR4 Q16900 Q296K0 D0V532 A0A182RSG8 Q16NR2 A0A182FS64 A0A1B0BSN1 W5JMH7 A0A182YHT8 A0A1A9Y3Z7 A0A1A9Z3Q5 A0A0L0BWF7 A0A195B8Y1 W8C0F5 T1EA28 A0A1L8DQG7 T1E7D1
W5JNQ1 A0A067RAJ4 E2AY08 E2C9W6 K7IV00 E2C9W4 A0A182TI71 A0A034WBG7 A0A0K8UWD9 E2C9W5 Q7PW17 A0A0K8VEN9 A0A0A1X0J3 A0A0K8U084 A0A195F7K8 A0A0C9S2B8 A0A0L0CPW6 A0A182VL28 A0A182R9M5 T1DPE8 A0A182QML4 A0A182TBT1 A0A182PPS1 A0A232EW13 A0A0C9RXJ7 E0VDU6 A0A084VG87 E5LCR7 B0W1J3 F4X0M1 W8BVD4 A0A182MMN1 D0R8R5 D2A5Z6 A0A232ET09 A0A158NDD6 A0A026WJK5 A0A151WYC0 A0A232EVX7 A0A0C9RRV3 K7IV02 A0A182J6W6 A0A0L7R675 D0R8R1 K7IZJ3 K7IV01 J9EB53 Q16H18 K7J1V4 E9IJV0 Q8T4T5 F4X0M2 B4NG84 A0A182X994 A0A084W0D6 A0A182PJ42 Q9BIG0 A0A1I8MJT5 Q16NR3 A0A0C9R9U5 B4GF17 A0A182QM63 A0A0L0CMG5 A0A1S4GBU1 A0A1I8P599 K7IV03 Q8N0R8 A0A3B0JP11 A0A182IMH4 A0A3L8DI96 A0A1L8DQU0 A0A182L8F6 A0A182GUH7 A0A1W4U884 Q7PX30 Q16NR4 Q16900 Q296K0 D0V532 A0A182RSG8 Q16NR2 A0A182FS64 A0A1B0BSN1 W5JMH7 A0A182YHT8 A0A1A9Y3Z7 A0A1A9Z3Q5 A0A0L0BWF7 A0A195B8Y1 W8C0F5 T1EA28 A0A1L8DQG7 T1E7D1
Pubmed
22118469
26354079
26227816
20920257
23761445
24845553
+ More
20798317 20075255 25348373 12364791 14747013 17210077 25830018 26108605 28648823 20566863 24438588 21699751 21719571 24495485 18362917 19820115 21347285 24508170 17510324 21282665 17994087 25315136 30249741 20966253 26483478 9134710 15632085 20017753 25244985
20798317 20075255 25348373 12364791 14747013 17210077 25830018 26108605 28648823 20566863 24438588 21699751 21719571 24495485 18362917 19820115 21347285 24508170 17510324 21282665 17994087 25315136 30249741 20966253 26483478 9134710 15632085 20017753 25244985
EMBL
AGBW02010309
OWR48755.1
NWSH01003453
PCG66270.1
ODYU01004678
SOQ44816.1
+ More
KQ459606 KPI91643.1 KQ461198 KPJ06235.1 JTDY01000047 KOB79190.1 ADMH02000581 ETN65771.1 KK852591 KDR20688.1 GL443740 EFN61711.1 GL453904 EFN75264.1 AAZX01009254 EFN75262.1 GAKP01007829 JAC51123.1 GDHF01021322 JAI30992.1 EFN75263.1 AAAB01008984 EAA14923.4 GDHF01015032 JAI37282.1 GBXI01009478 JAD04814.1 GDHF01032378 JAI19936.1 KQ981744 KYN36366.1 GBYB01015056 JAG84823.1 JRES01000186 KNC33479.1 GAMD01003399 JAA98191.1 AXCN02000839 NNAY01001939 OXU22501.1 GBYB01013885 JAG83652.1 DS235088 EEB11552.1 ATLV01012699 KE524808 KFB36981.1 HQ424574 ADP88998.1 DS231822 EDS25791.1 GL888498 EGI60013.1 GAMC01001265 JAC05291.1 AXCM01001573 FN547064 CBC01179.1 KQ971345 EFA05680.2 NNAY01002348 OXU21493.1 ADTU01012123 ADTU01012124 KK107213 EZA55304.1 KQ982650 KYQ52884.1 OXU22502.1 GBYB01011270 JAG81037.1 KQ414646 KOC66382.1 FN547060 KQ971361 CBC01171.1 EFA05699.1 CH477811 EJY57946.1 CH478212 EAT33546.1 GL763883 EFZ19174.1 AY064082 AAL85583.1 EGI60014.1 CH964251 EDW83301.1 ATLV01019117 KE525262 KFB43680.1 AY038040 AAK72504.1 EAT35997.1 EJY57945.1 GBYB01003581 JAG73348.1 CH479182 EDW34202.1 AXCN02001492 KNC33490.1 AAAB01008987 AY008349 AAK13194.3 OUUW01000008 SPP83865.1 QOIP01000007 RLU20046.1 GFDF01005374 JAV08710.1 JXUM01089189 KQ563751 KXJ73382.1 EAA00961.5 EAT35995.1 U56423 AY008348 AY038039 AY064083 CH477261 AAB01218.1 AAK13193.1 AAK72503.1 AAL85584.1 EAT45655.1 CM000070 EAL28457.1 GU017991 ACY24330.1 EAT35998.1 JXJN01019786 ADMH02001098 ETN64110.1 JRES01001243 KNC24362.1 KQ976558 KYM80690.1 GAMC01001408 JAC05148.1 GAMD01001378 JAB00213.1 GFDF01005381 JAV08703.1 GAMD01003147 JAA98443.1
KQ459606 KPI91643.1 KQ461198 KPJ06235.1 JTDY01000047 KOB79190.1 ADMH02000581 ETN65771.1 KK852591 KDR20688.1 GL443740 EFN61711.1 GL453904 EFN75264.1 AAZX01009254 EFN75262.1 GAKP01007829 JAC51123.1 GDHF01021322 JAI30992.1 EFN75263.1 AAAB01008984 EAA14923.4 GDHF01015032 JAI37282.1 GBXI01009478 JAD04814.1 GDHF01032378 JAI19936.1 KQ981744 KYN36366.1 GBYB01015056 JAG84823.1 JRES01000186 KNC33479.1 GAMD01003399 JAA98191.1 AXCN02000839 NNAY01001939 OXU22501.1 GBYB01013885 JAG83652.1 DS235088 EEB11552.1 ATLV01012699 KE524808 KFB36981.1 HQ424574 ADP88998.1 DS231822 EDS25791.1 GL888498 EGI60013.1 GAMC01001265 JAC05291.1 AXCM01001573 FN547064 CBC01179.1 KQ971345 EFA05680.2 NNAY01002348 OXU21493.1 ADTU01012123 ADTU01012124 KK107213 EZA55304.1 KQ982650 KYQ52884.1 OXU22502.1 GBYB01011270 JAG81037.1 KQ414646 KOC66382.1 FN547060 KQ971361 CBC01171.1 EFA05699.1 CH477811 EJY57946.1 CH478212 EAT33546.1 GL763883 EFZ19174.1 AY064082 AAL85583.1 EGI60014.1 CH964251 EDW83301.1 ATLV01019117 KE525262 KFB43680.1 AY038040 AAK72504.1 EAT35997.1 EJY57945.1 GBYB01003581 JAG73348.1 CH479182 EDW34202.1 AXCN02001492 KNC33490.1 AAAB01008987 AY008349 AAK13194.3 OUUW01000008 SPP83865.1 QOIP01000007 RLU20046.1 GFDF01005374 JAV08710.1 JXUM01089189 KQ563751 KXJ73382.1 EAA00961.5 EAT35995.1 U56423 AY008348 AY038039 AY064083 CH477261 AAB01218.1 AAK13193.1 AAK72503.1 AAL85584.1 EAT45655.1 CM000070 EAL28457.1 GU017991 ACY24330.1 EAT35998.1 JXJN01019786 ADMH02001098 ETN64110.1 JRES01001243 KNC24362.1 KQ976558 KYM80690.1 GAMC01001408 JAC05148.1 GAMD01001378 JAB00213.1 GFDF01005381 JAV08703.1 GAMD01003147 JAA98443.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
UP000000673
+ More
UP000027135 UP000000311 UP000008237 UP000002358 UP000075902 UP000007062 UP000078541 UP000037069 UP000075903 UP000075900 UP000075886 UP000075901 UP000075885 UP000215335 UP000009046 UP000030765 UP000002320 UP000007755 UP000075883 UP000007266 UP000005205 UP000053097 UP000075809 UP000075880 UP000053825 UP000008820 UP000007798 UP000076407 UP000095301 UP000008744 UP000095300 UP000268350 UP000279307 UP000075882 UP000069940 UP000249989 UP000192221 UP000001819 UP000069272 UP000092460 UP000076408 UP000092443 UP000092445 UP000078540
UP000027135 UP000000311 UP000008237 UP000002358 UP000075902 UP000007062 UP000078541 UP000037069 UP000075903 UP000075900 UP000075886 UP000075901 UP000075885 UP000215335 UP000009046 UP000030765 UP000002320 UP000007755 UP000075883 UP000007266 UP000005205 UP000053097 UP000075809 UP000075880 UP000053825 UP000008820 UP000007798 UP000076407 UP000095301 UP000008744 UP000095300 UP000268350 UP000279307 UP000075882 UP000069940 UP000249989 UP000192221 UP000001819 UP000069272 UP000092460 UP000076408 UP000092443 UP000092445 UP000078540
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A212F4T3
A0A2A4J497
A0A2H1VVN5
A0A194PF43
A0A194QMP7
A0A0L7LUS5
+ More
W5JNQ1 A0A067RAJ4 E2AY08 E2C9W6 K7IV00 E2C9W4 A0A182TI71 A0A034WBG7 A0A0K8UWD9 E2C9W5 Q7PW17 A0A0K8VEN9 A0A0A1X0J3 A0A0K8U084 A0A195F7K8 A0A0C9S2B8 A0A0L0CPW6 A0A182VL28 A0A182R9M5 T1DPE8 A0A182QML4 A0A182TBT1 A0A182PPS1 A0A232EW13 A0A0C9RXJ7 E0VDU6 A0A084VG87 E5LCR7 B0W1J3 F4X0M1 W8BVD4 A0A182MMN1 D0R8R5 D2A5Z6 A0A232ET09 A0A158NDD6 A0A026WJK5 A0A151WYC0 A0A232EVX7 A0A0C9RRV3 K7IV02 A0A182J6W6 A0A0L7R675 D0R8R1 K7IZJ3 K7IV01 J9EB53 Q16H18 K7J1V4 E9IJV0 Q8T4T5 F4X0M2 B4NG84 A0A182X994 A0A084W0D6 A0A182PJ42 Q9BIG0 A0A1I8MJT5 Q16NR3 A0A0C9R9U5 B4GF17 A0A182QM63 A0A0L0CMG5 A0A1S4GBU1 A0A1I8P599 K7IV03 Q8N0R8 A0A3B0JP11 A0A182IMH4 A0A3L8DI96 A0A1L8DQU0 A0A182L8F6 A0A182GUH7 A0A1W4U884 Q7PX30 Q16NR4 Q16900 Q296K0 D0V532 A0A182RSG8 Q16NR2 A0A182FS64 A0A1B0BSN1 W5JMH7 A0A182YHT8 A0A1A9Y3Z7 A0A1A9Z3Q5 A0A0L0BWF7 A0A195B8Y1 W8C0F5 T1EA28 A0A1L8DQG7 T1E7D1
W5JNQ1 A0A067RAJ4 E2AY08 E2C9W6 K7IV00 E2C9W4 A0A182TI71 A0A034WBG7 A0A0K8UWD9 E2C9W5 Q7PW17 A0A0K8VEN9 A0A0A1X0J3 A0A0K8U084 A0A195F7K8 A0A0C9S2B8 A0A0L0CPW6 A0A182VL28 A0A182R9M5 T1DPE8 A0A182QML4 A0A182TBT1 A0A182PPS1 A0A232EW13 A0A0C9RXJ7 E0VDU6 A0A084VG87 E5LCR7 B0W1J3 F4X0M1 W8BVD4 A0A182MMN1 D0R8R5 D2A5Z6 A0A232ET09 A0A158NDD6 A0A026WJK5 A0A151WYC0 A0A232EVX7 A0A0C9RRV3 K7IV02 A0A182J6W6 A0A0L7R675 D0R8R1 K7IZJ3 K7IV01 J9EB53 Q16H18 K7J1V4 E9IJV0 Q8T4T5 F4X0M2 B4NG84 A0A182X994 A0A084W0D6 A0A182PJ42 Q9BIG0 A0A1I8MJT5 Q16NR3 A0A0C9R9U5 B4GF17 A0A182QM63 A0A0L0CMG5 A0A1S4GBU1 A0A1I8P599 K7IV03 Q8N0R8 A0A3B0JP11 A0A182IMH4 A0A3L8DI96 A0A1L8DQU0 A0A182L8F6 A0A182GUH7 A0A1W4U884 Q7PX30 Q16NR4 Q16900 Q296K0 D0V532 A0A182RSG8 Q16NR2 A0A182FS64 A0A1B0BSN1 W5JMH7 A0A182YHT8 A0A1A9Y3Z7 A0A1A9Z3Q5 A0A0L0BWF7 A0A195B8Y1 W8C0F5 T1EA28 A0A1L8DQG7 T1E7D1
PDB
4BNR
E-value=2.31591e-29,
Score=319
Ontologies
GO
Topology
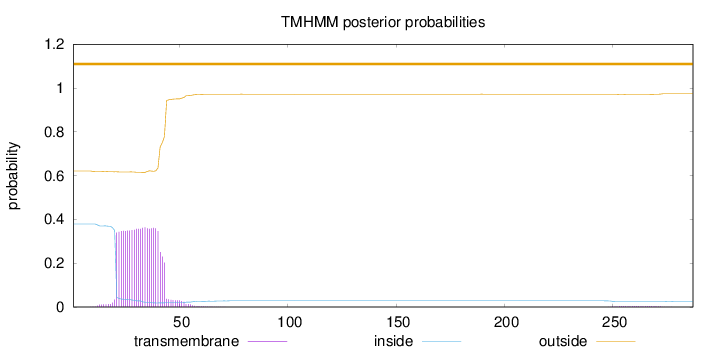
Length:
287
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.32968999999997
Exp number, first 60 AAs:
8.22243
Total prob of N-in:
0.37933
outside
1 - 287
Population Genetic Test Statistics
Pi
191.223086
Theta
179.155617
Tajima's D
0.641326
CLR
0
CSRT
0.555272236388181
Interpretation
Uncertain
