Gene
KWMTBOMO05553 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006745
Annotation
laccase_2A_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.745
Sequence
CDS
ATGAAACAGACCGAAACGTTCAACCTTGTACTCCAGCTTAATACTTTTTTGCCTTTGATTTCAGAAGCCATCAACGCCGCAGACGACCAGTCCACATCGGCATCGTGGTGGCAAGCCGGGACAGCAACACCGTTCAGGGAAACGGGCAGTAATGCGTTCTCATCCACCCATGGACTCGTTCAAACACACCCCAGCGCTGATGATCCATTCGGAACTACTTTTGGAGGAATAGGAAGTACTATAGGACCGTCATCGAATCCCTTCGCTAAAAGCGGCAGCGGTCCGCTGAGTGGAGGCGTTAGGAAGAATCCTTTGCCATCTCTAGCCAGGATCTCTAACATTAACGGAAAAGCCTCATTGAAACATTTGGATTTTACTAGTAGTGCTACCGCTGAATTGAGAAGGAACCCAGCACTGTCGGCCCCTGATGAGTGTGCCAGAGCCTGCAGGGAAAACGAACCACCAAGGATCTGTTACTATCATTTCACATTGGAGTTTTATACTGTCTTGGGAGCGGCTTGTCAAGTATGCACACCAAACGCTACGAATGTGGTCTGGTCTCATTGCCAATGCGTACTGGCCGATGGCGTCGAACGGGGTATTCTCACTGCCAACAGAATGTTACCAGGCCCTTCCATTCAAGTTTGCGAAAACGATAAAGTGGTAATTGACGTAGAGAACCATATGGAAGGTATGGAGGTGACAATCCATTGGCACGGTATTTGGCAAAGGGGATCACAATATTACGATGGTGTACCGTTTGTCACGCAATGTCCTATTCAGCAAGGGAATACTTTCAGATATCAATGGCAAGGTAATGCTGGCACTCACTTCTGGCACGCTCACACTGGACTACAGAAACTGGATGGTTTATACGGCAGCATTGTCGTTCGTCAACCTCCTTCAAAAGATCCCAACAGTCACTTATACGATTACGATTTGACCACACACGTCATGTTGATCAGTGACTGGCTCCACGAAGATGCCGCTGAAAGATACCCAGGACGTTTAGCTGTCAATACTGGACAGGACCCCGAATCTGTATTGATCAACGGAAAAGGACAATTCAGAGATCCTAACACTGGCTTCATGACCAATACTCCATTGGAAGTTTTCACAATTACTCCTGGTAGACGTTACAGGTTCAGAATGATCAATGCCTTTGCATCGGTATGCCCTGCTCAAATCACGTTTGAAGGTCACAACTTGACAGTTATTGCTACTGACGGTGAGCCAGTGCAACCCGTGCAAGTGAACACGATAATCTCGTTCTCAGGGGAACGATACGACTTTGTAATCGAAGCTAACAACATTCCCGGAGCATACTGGATCCAAGTTCGAGGTCTTGGTGAATGTGGTATTAAGAGAGCACAACAATTAGGTATCCTCAGATACGCTAGAGGACCTTACCAACCATCTTCGCTGGCCCCCACCTACGATGTCGGAATACCCCAAGGAGTGGTGATGAACCCCTTGGACGCGAGATGTAATATATCAAGAAACGATGCTATTTGCGTGAGCCAATTGAAAAACGCTCAGAACATCGACCCCGCCATCCTTCAGGAGAGACCCGACGTTAAAATCTTTTTGCCCTTCCGATTCTTCGTATACAGACCAGAGATGCTCTTCCAACCCAACACATACAACAGATACTTGGTTGCCCCTGGAGGAGACCACGTTATCAGTTTGATTGACGAAATCTCATATATGTCCCCGCCTGCTCCATTGATCAGTCAGTATGATGACATCAATCCTGACCAATTCTGTAATGGAGACAACCGACCCGCTAACTGTGGACAAAACTGCATGTGCACACACAAAGTTGACATACCTCTGAATGCTGTTGTGGAGATTGTATTGGTAGATGAAGTACAAATCGCGAACTTATCTCATCCGTTCCACTTGCACGGATATTCATACAACGTGATAGGTATCGGAAGATCACCCGACCAGAACGTTAAGAAAATTAACTTGAAGCACGCACTCGACCTCGACAGAAGAGGATTGCTCGAACGTCACCTGAAGCAAGGAGACCTTCCTCCTGCAAAAGACACCATCGCTGTTCCCAACAACGGCTACGTCATTCTCCGTTTCAGAGCCACAAATCCTGGCTTCTGGCTACTCCATTGCCACTTCCTTTTCCACATCGTCATCGGAATGAGCTTGGTGCTCCAAGTAGGCACGCAGGGCGACCTTCCCCCCGTACCCCCGAACTTCCCCACATGCGGAGACCATCTCCCAGCCATCCCACTCCACTGA
Protein
MKQTETFNLVLQLNTFLPLISEAINAADDQSTSASWWQAGTATPFRETGSNAFSSTHGLVQTHPSADDPFGTTFGGIGSTIGPSSNPFAKSGSGPLSGGVRKNPLPSLARISNINGKASLKHLDFTSSATAELRRNPALSAPDECARACRENEPPRICYYHFTLEFYTVLGAACQVCTPNATNVVWSHCQCVLADGVERGILTANRMLPGPSIQVCENDKVVIDVENHMEGMEVTIHWHGIWQRGSQYYDGVPFVTQCPIQQGNTFRYQWQGNAGTHFWHAHTGLQKLDGLYGSIVVRQPPSKDPNSHLYDYDLTTHVMLISDWLHEDAAERYPGRLAVNTGQDPESVLINGKGQFRDPNTGFMTNTPLEVFTITPGRRYRFRMINAFASVCPAQITFEGHNLTVIATDGEPVQPVQVNTIISFSGERYDFVIEANNIPGAYWIQVRGLGECGIKRAQQLGILRYARGPYQPSSLAPTYDVGIPQGVVMNPLDARCNISRNDAICVSQLKNAQNIDPAILQERPDVKIFLPFRFFVYRPEMLFQPNTYNRYLVAPGGDHVISLIDEISYMSPPAPLISQYDDINPDQFCNGDNRPANCGQNCMCTHKVDIPLNAVVEIVLVDEVQIANLSHPFHLHGYSYNVIGIGRSPDQNVKKINLKHALDLDRRGLLERHLKQGDLPPAKDTIAVPNNGYVILRFRATNPGFWLLHCHFLFHIVIGMSLVLQVGTQGDLPPVPPNFPTCGDHLPAIPLH
Summary
Similarity
Belongs to the multicopper oxidase family.
Uniprot
B5BR55
A7XQR9
H9JB50
A0A076FLX5
Q8I8Y0
A0A212F4T2
+ More
D5MRR9 D4AH59 A0A2H1VVE5 A0A2A4J3K6 D5MRS1 U6A581 B4F7L6 A0A194PG16 A0A0L7LUX0 G4XHA2 A0A084WL27 A0A1W5VPG6 A0A182LS40 A0A182IZ20 Q4ZGM4 A0A182UT19 A7UU38 A0A182I1A0 Q58IU3 B4KLT3 B5B2D0 B4GBH6 A0A0J9R682 A0A0J9R6H4 B4IM24 B4LPG0 B0XBY5 B0WW28 Q7JQF6 A0A182P7P1 A1Z6F6 Q29CP8 B3N3L4 B3N1H7 B4ISZ0 A0A182YKV7 A0A182X5E6 A0A0P8XD20 A0A336LNA3 A0A182RDU7 Q86P50 A0A182WLN0 A0A1I8MAX1 A0A182U114 A0A1I8QC41 A0A034V864 T1PG09 A0A0A1WT11 W8BXM7 A0A182NK10 A0A0L0CHR0 A0A182KNW0 B4J6G7 A0A0K8VT75 A0A194QL27 A0A182FHY6 A0A0P6IU09 Q0IF14 Q58IU2 Q7Q5V9 A0A0J9R7F0 D6WE75 A0A1A9ZUU7 Q49I41 A1Z6F4 A0A1W4WWP4 B7YZT1 A0A0J9R5Z8 A0A2U8ICW4 A0A1Y1MUB4 A0A0J9TTT8 A0A1B0AMG9 A0A0B4KF30 A0A182QNY9 A0A2U8ICP2 A0A1A9YJF8 A0A0Q5WLN4 Q49I40 A0A2M4CZW5 W5JD72 A7XQS2 A0A087ZPF2 B4MQV3 A0A034V974 A0A0N0BD48 A0A1J1J067 A0A3L8DUA5 A0A0C9PH46 A0A154PKC4 B4K2F0 E9I908 A0A2A3EF34 A0A195ATS4 A0A0L7R085
D5MRR9 D4AH59 A0A2H1VVE5 A0A2A4J3K6 D5MRS1 U6A581 B4F7L6 A0A194PG16 A0A0L7LUX0 G4XHA2 A0A084WL27 A0A1W5VPG6 A0A182LS40 A0A182IZ20 Q4ZGM4 A0A182UT19 A7UU38 A0A182I1A0 Q58IU3 B4KLT3 B5B2D0 B4GBH6 A0A0J9R682 A0A0J9R6H4 B4IM24 B4LPG0 B0XBY5 B0WW28 Q7JQF6 A0A182P7P1 A1Z6F6 Q29CP8 B3N3L4 B3N1H7 B4ISZ0 A0A182YKV7 A0A182X5E6 A0A0P8XD20 A0A336LNA3 A0A182RDU7 Q86P50 A0A182WLN0 A0A1I8MAX1 A0A182U114 A0A1I8QC41 A0A034V864 T1PG09 A0A0A1WT11 W8BXM7 A0A182NK10 A0A0L0CHR0 A0A182KNW0 B4J6G7 A0A0K8VT75 A0A194QL27 A0A182FHY6 A0A0P6IU09 Q0IF14 Q58IU2 Q7Q5V9 A0A0J9R7F0 D6WE75 A0A1A9ZUU7 Q49I41 A1Z6F4 A0A1W4WWP4 B7YZT1 A0A0J9R5Z8 A0A2U8ICW4 A0A1Y1MUB4 A0A0J9TTT8 A0A1B0AMG9 A0A0B4KF30 A0A182QNY9 A0A2U8ICP2 A0A1A9YJF8 A0A0Q5WLN4 Q49I40 A0A2M4CZW5 W5JD72 A7XQS2 A0A087ZPF2 B4MQV3 A0A034V974 A0A0N0BD48 A0A1J1J067 A0A3L8DUA5 A0A0C9PH46 A0A154PKC4 B4K2F0 E9I908 A0A2A3EF34 A0A195ATS4 A0A0L7R085
Pubmed
14614147
19168135
18477244
19121390
22118469
26354079
+ More
26227816 24438588 28376914 12364791 14747013 17210077 18675911 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 25244985 25315136 25348373 25830018 24495485 26108605 20966253 26999592 17510324 18362917 19820115 16076951 28004739 20920257 23761445 30249741 21282665
26227816 24438588 28376914 12364791 14747013 17210077 18675911 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 25244985 25315136 25348373 25830018 24495485 26108605 20966253 26999592 17510324 18362917 19820115 16076951 28004739 20920257 23761445 30249741 21282665
EMBL
AB379590
BAG70891.1
EU093074
ABU68465.1
BABH01008618
KJ789368
+ More
AII19522.1 AY135186 AAN17507.1 AGBW02010309 OWR48758.1 AB531133 BAJ07600.1 AB499123 BAI87829.1 ODYU01004678 SOQ44817.1 NWSH01003453 PCG66268.1 AB531135 BAJ07602.1 KF479388 AHA15412.1 BK006378 DAA06287.1 KQ459606 KPI91644.1 JTDY01000047 KOB79189.1 JF811457 AEP43806.1 ATLV01024176 KE525350 KFB50921.1 KY132102 ARG47519.1 AXCM01006745 DQ011288 AAY29698.1 AAAB01008960 EDO63816.1 APCN01000047 APCN01000048 AY943928 AAX49501.1 CH933808 EDW09743.1 EU886786 ACG63789.1 CH479181 EDW31271.1 CM002911 KMY91618.1 KMY91621.1 CH480920 EDW43632.1 CH940648 EDW61219.2 DS232661 EDS44514.1 DS232136 EDS35860.1 AE013599 BT010091 AAF57332.4 AAQ22560.1 BT133179 AAF57331.3 AEZ68807.1 CH475437 EAL29272.3 CH954177 EDV59896.1 CH902656 EDV29979.2 CH891608 EDW99572.1 KPU72643.1 UFQT01000078 SSX19430.1 BT003483 AAO39486.1 GAKP01020651 GAKP01020650 JAC38302.1 KA647757 AFP62386.1 GBXI01012310 GBXI01009830 GBXI01008054 GBXI01005511 JAD01982.1 JAD04462.1 JAD06238.1 JAD08781.1 GAMC01012586 GAMC01012585 JAB93969.1 JRES01000379 KNC31786.1 CH916367 EDW01968.1 GDHF01020306 GDHF01013149 GDHF01012944 GDHF01010584 GDHF01009008 JAI32008.1 JAI39165.1 JAI39370.1 JAI41730.1 JAI43306.1 KQ461198 KPJ06237.1 GDUN01000603 JAN95316.1 CH477440 EAT40867.1 AY943929 AAX49502.1 EAA11473.4 KMY91619.1 KQ971324 EFA01166.1 AY884061 AAX84202.2 AAN16124.2 ACL83060.1 KMY91622.1 MH253688 AWK23446.1 GEZM01020491 JAV89262.1 KMY91620.1 JXJN01000432 JXJN01000433 JXJN01000434 JXJN01000435 AGB93244.1 AXCN02000495 MH253687 AWK23445.1 KQS71051.1 AY884062 AAX84203.2 GGFL01006686 MBW70864.1 ADMH02001577 ETN62011.1 EU093075 ABU68466.1 CH963849 EDW74492.1 GAKP01020652 JAC38300.1 KQ435878 KOX69942.1 CVRI01000066 CRL05915.1 QOIP01000004 RLU24055.1 GBYB01000178 JAG69945.1 KQ434943 KZC12283.1 CH919043 EDW05139.1 GL761667 EFZ22928.1 KZ288262 PBC30347.1 KQ976741 KYM75596.1 KQ414672 KOC64253.1
AII19522.1 AY135186 AAN17507.1 AGBW02010309 OWR48758.1 AB531133 BAJ07600.1 AB499123 BAI87829.1 ODYU01004678 SOQ44817.1 NWSH01003453 PCG66268.1 AB531135 BAJ07602.1 KF479388 AHA15412.1 BK006378 DAA06287.1 KQ459606 KPI91644.1 JTDY01000047 KOB79189.1 JF811457 AEP43806.1 ATLV01024176 KE525350 KFB50921.1 KY132102 ARG47519.1 AXCM01006745 DQ011288 AAY29698.1 AAAB01008960 EDO63816.1 APCN01000047 APCN01000048 AY943928 AAX49501.1 CH933808 EDW09743.1 EU886786 ACG63789.1 CH479181 EDW31271.1 CM002911 KMY91618.1 KMY91621.1 CH480920 EDW43632.1 CH940648 EDW61219.2 DS232661 EDS44514.1 DS232136 EDS35860.1 AE013599 BT010091 AAF57332.4 AAQ22560.1 BT133179 AAF57331.3 AEZ68807.1 CH475437 EAL29272.3 CH954177 EDV59896.1 CH902656 EDV29979.2 CH891608 EDW99572.1 KPU72643.1 UFQT01000078 SSX19430.1 BT003483 AAO39486.1 GAKP01020651 GAKP01020650 JAC38302.1 KA647757 AFP62386.1 GBXI01012310 GBXI01009830 GBXI01008054 GBXI01005511 JAD01982.1 JAD04462.1 JAD06238.1 JAD08781.1 GAMC01012586 GAMC01012585 JAB93969.1 JRES01000379 KNC31786.1 CH916367 EDW01968.1 GDHF01020306 GDHF01013149 GDHF01012944 GDHF01010584 GDHF01009008 JAI32008.1 JAI39165.1 JAI39370.1 JAI41730.1 JAI43306.1 KQ461198 KPJ06237.1 GDUN01000603 JAN95316.1 CH477440 EAT40867.1 AY943929 AAX49502.1 EAA11473.4 KMY91619.1 KQ971324 EFA01166.1 AY884061 AAX84202.2 AAN16124.2 ACL83060.1 KMY91622.1 MH253688 AWK23446.1 GEZM01020491 JAV89262.1 KMY91620.1 JXJN01000432 JXJN01000433 JXJN01000434 JXJN01000435 AGB93244.1 AXCN02000495 MH253687 AWK23445.1 KQS71051.1 AY884062 AAX84203.2 GGFL01006686 MBW70864.1 ADMH02001577 ETN62011.1 EU093075 ABU68466.1 CH963849 EDW74492.1 GAKP01020652 JAC38300.1 KQ435878 KOX69942.1 CVRI01000066 CRL05915.1 QOIP01000004 RLU24055.1 GBYB01000178 JAG69945.1 KQ434943 KZC12283.1 CH919043 EDW05139.1 GL761667 EFZ22928.1 KZ288262 PBC30347.1 KQ976741 KYM75596.1 KQ414672 KOC64253.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000037510
UP000030765
+ More
UP000075883 UP000075880 UP000075903 UP000007062 UP000075840 UP000009192 UP000008744 UP000001292 UP000008792 UP000002320 UP000000803 UP000075885 UP000001819 UP000008711 UP000007801 UP000002282 UP000076408 UP000076407 UP000075900 UP000075920 UP000095301 UP000075902 UP000095300 UP000075884 UP000037069 UP000075882 UP000001070 UP000053240 UP000069272 UP000008820 UP000007266 UP000092445 UP000192223 UP000092460 UP000075886 UP000092443 UP000000673 UP000005203 UP000007798 UP000053105 UP000183832 UP000279307 UP000076502 UP000242457 UP000078540 UP000053825
UP000075883 UP000075880 UP000075903 UP000007062 UP000075840 UP000009192 UP000008744 UP000001292 UP000008792 UP000002320 UP000000803 UP000075885 UP000001819 UP000008711 UP000007801 UP000002282 UP000076408 UP000076407 UP000075900 UP000075920 UP000095301 UP000075902 UP000095300 UP000075884 UP000037069 UP000075882 UP000001070 UP000053240 UP000069272 UP000008820 UP000007266 UP000092445 UP000192223 UP000092460 UP000075886 UP000092443 UP000000673 UP000005203 UP000007798 UP000053105 UP000183832 UP000279307 UP000076502 UP000242457 UP000078540 UP000053825
Interpro
SUPFAM
SSF49503
SSF49503
Gene 3D
ProteinModelPortal
B5BR55
A7XQR9
H9JB50
A0A076FLX5
Q8I8Y0
A0A212F4T2
+ More
D5MRR9 D4AH59 A0A2H1VVE5 A0A2A4J3K6 D5MRS1 U6A581 B4F7L6 A0A194PG16 A0A0L7LUX0 G4XHA2 A0A084WL27 A0A1W5VPG6 A0A182LS40 A0A182IZ20 Q4ZGM4 A0A182UT19 A7UU38 A0A182I1A0 Q58IU3 B4KLT3 B5B2D0 B4GBH6 A0A0J9R682 A0A0J9R6H4 B4IM24 B4LPG0 B0XBY5 B0WW28 Q7JQF6 A0A182P7P1 A1Z6F6 Q29CP8 B3N3L4 B3N1H7 B4ISZ0 A0A182YKV7 A0A182X5E6 A0A0P8XD20 A0A336LNA3 A0A182RDU7 Q86P50 A0A182WLN0 A0A1I8MAX1 A0A182U114 A0A1I8QC41 A0A034V864 T1PG09 A0A0A1WT11 W8BXM7 A0A182NK10 A0A0L0CHR0 A0A182KNW0 B4J6G7 A0A0K8VT75 A0A194QL27 A0A182FHY6 A0A0P6IU09 Q0IF14 Q58IU2 Q7Q5V9 A0A0J9R7F0 D6WE75 A0A1A9ZUU7 Q49I41 A1Z6F4 A0A1W4WWP4 B7YZT1 A0A0J9R5Z8 A0A2U8ICW4 A0A1Y1MUB4 A0A0J9TTT8 A0A1B0AMG9 A0A0B4KF30 A0A182QNY9 A0A2U8ICP2 A0A1A9YJF8 A0A0Q5WLN4 Q49I40 A0A2M4CZW5 W5JD72 A7XQS2 A0A087ZPF2 B4MQV3 A0A034V974 A0A0N0BD48 A0A1J1J067 A0A3L8DUA5 A0A0C9PH46 A0A154PKC4 B4K2F0 E9I908 A0A2A3EF34 A0A195ATS4 A0A0L7R085
D5MRR9 D4AH59 A0A2H1VVE5 A0A2A4J3K6 D5MRS1 U6A581 B4F7L6 A0A194PG16 A0A0L7LUX0 G4XHA2 A0A084WL27 A0A1W5VPG6 A0A182LS40 A0A182IZ20 Q4ZGM4 A0A182UT19 A7UU38 A0A182I1A0 Q58IU3 B4KLT3 B5B2D0 B4GBH6 A0A0J9R682 A0A0J9R6H4 B4IM24 B4LPG0 B0XBY5 B0WW28 Q7JQF6 A0A182P7P1 A1Z6F6 Q29CP8 B3N3L4 B3N1H7 B4ISZ0 A0A182YKV7 A0A182X5E6 A0A0P8XD20 A0A336LNA3 A0A182RDU7 Q86P50 A0A182WLN0 A0A1I8MAX1 A0A182U114 A0A1I8QC41 A0A034V864 T1PG09 A0A0A1WT11 W8BXM7 A0A182NK10 A0A0L0CHR0 A0A182KNW0 B4J6G7 A0A0K8VT75 A0A194QL27 A0A182FHY6 A0A0P6IU09 Q0IF14 Q58IU2 Q7Q5V9 A0A0J9R7F0 D6WE75 A0A1A9ZUU7 Q49I41 A1Z6F4 A0A1W4WWP4 B7YZT1 A0A0J9R5Z8 A0A2U8ICW4 A0A1Y1MUB4 A0A0J9TTT8 A0A1B0AMG9 A0A0B4KF30 A0A182QNY9 A0A2U8ICP2 A0A1A9YJF8 A0A0Q5WLN4 Q49I40 A0A2M4CZW5 W5JD72 A7XQS2 A0A087ZPF2 B4MQV3 A0A034V974 A0A0N0BD48 A0A1J1J067 A0A3L8DUA5 A0A0C9PH46 A0A154PKC4 B4K2F0 E9I908 A0A2A3EF34 A0A195ATS4 A0A0L7R085
PDB
5MIG
E-value=8.7223e-47,
Score=474
Ontologies
GO
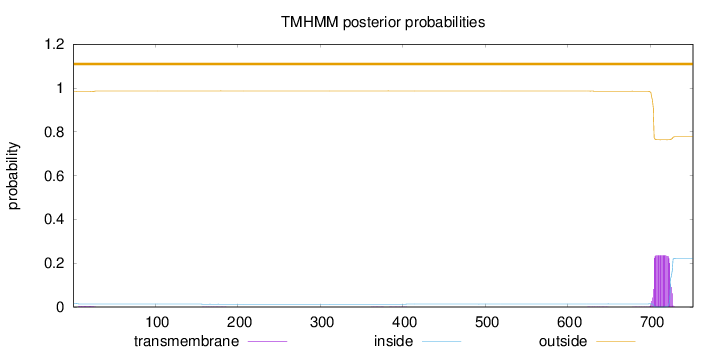
Topology
Length:
752
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.176
Exp number, first 60 AAs:
0.05991
Total prob of N-in:
0.01659
outside
1 - 752
Population Genetic Test Statistics
Pi
286.796102
Theta
228.00198
Tajima's D
0.86703
CLR
0.48847
CSRT
0.618269086545673
Interpretation
Uncertain
