Gene
KWMTBOMO05547
Pre Gene Modal
BGIBMGA006601
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_70_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.385 Nuclear Reliability : 1.442
Sequence
CDS
ATGTGGCCTCGCTGGTTGTTATTATGCTTGGTGGTGCTATACGCGCGGCCAGTACACCCACAAGGCGCCCAGCAGTGCCCCGCTCCGAACACCATGACACCTTGCAGCTGCACGGTCAAGAAAAACGGCTTGGATATACTCTGCGAGTTCACTGAACAGCAACATATACAAAAGGCAATGAACACGCTGCGTTCGAAGCCAATGATTATATTTTATCTGAAGCTACGTCACAACAACCTGGCTAAGTTGCCCTCTTACATATTCCTGGGGCTCGATATACGTCATCTCACTGTTCACAACAGCAGCCTAGCGGTCATCGAGGATTCTTCACTAAGCTCCATCGGCAACAAGCTGACTCAGCTGGACGTCTCCCAGAATAGTCTGGCCACAATACCGACATCTTCGTTCACGCATCTCAACCACCTGCTCATACTGAACATGAATCATAACAAGGTGACCGCGGTACACAACAGAGCATTCGTCGGACTTGATACCCTAGAGATACTGACTCTGTATGAAAATAGGATTTCGGTAGTCGATGGCGAGGCTTTCAAAGGATTGGAAAAGAAGCTTAAGCGATTAAATCTTGGAGGCAATGAATTGACGGCGGTCCCGCAAAAAGCGCTGGCTCTTCTCGAGAATTTGAAAAAACTTGAAATGCAAGAAAATCGGATTACTTCTATTTCAGAAGGGGACTTTGCAGGGTTACGTAACCTAGATTCTTTAGGCCTAGCTCATAACCAGCTTCGCGAAGTTCCGGCTCAAGTGTTTTCGCACCTTACGTTTTTGAACTCACTGGAGTTGGAGGGAAACCTGATTCAACGTATTGACGAGAAAGCCTTTGCTGGACTTGAAGAAAATCTGCAATACTTACGTCTTGGCGACAACCGCCTACATGAGATCCCCTCTGAAGCACTGCGTCCTCTGCATCGTCTCCGCCATCTGGACCTTCGATCCAACAACATAACATACATCAGTGAAGATGCCTTCACTGGCTACGGAGACTCCATCACGTTCCTCAACATGCAAAAGAATATGATTCATCAGCTGCCAACAATGGGCTTCGATAATCTGAACTCCCTGGAAACACTGAACCTGCAAAACAATAAACTACAGCATATTCCTGAGGAAATAATGGAGCCGATACTCGATACATTACGAGTCGTGGATATAATGGATAATCCTCTGATCTGTGACTGTGAGCTGGCTTGGTACGAAGCTTGGTTGGCCGGACTCCGCGACCGCGACGATGAGATGATGCAGAAGAAACGGACAGTATGTACCATGCTCAACGAGCATCGCGAATACAGCGTCTCTAAAATGCCGCTCGAAAAGATGAGCTGCAAAAGAAAACCCACCTACGGCCGAACATCGGGTGCGGATACACTCAAAAAATTCCGGGCCACCATGATCTGTAAGCTAGTTTTACCGTAA
Protein
MWPRWLLLCLVVLYARPVHPQGAQQCPAPNTMTPCSCTVKKNGLDILCEFTEQQHIQKAMNTLRSKPMIIFYLKLRHNNLAKLPSYIFLGLDIRHLTVHNSSLAVIEDSSLSSIGNKLTQLDVSQNSLATIPTSSFTHLNHLLILNMNHNKVTAVHNRAFVGLDTLEILTLYENRISVVDGEAFKGLEKKLKRLNLGGNELTAVPQKALALLENLKKLEMQENRITSISEGDFAGLRNLDSLGLAHNQLREVPAQVFSHLTFLNSLELEGNLIQRIDEKAFAGLEENLQYLRLGDNRLHEIPSEALRPLHRLRHLDLRSNNITYISEDAFTGYGDSITFLNMQKNMIHQLPTMGFDNLNSLETLNLQNNKLQHIPEEIMEPILDTLRVVDIMDNPLICDCELAWYEAWLAGLRDRDDEMMQKKRTVCTMLNEHREYSVSKMPLEKMSCKRKPTYGRTSGADTLKKFRATMICKLVLP
Summary
Uniprot
A0A2A4K690
A0A212ES11
H9JAQ6
A0A194PEF0
A0A0L7LVM3
A0A194QMQ2
+ More
A0A1B6LR65 A0A1B6GGD2 A0A1B6GL60 A0A1B6DK70 U4U2K4 N6UFH3 A0A139WCJ5 A0A1Y1LFZ3 A0A026X1V6 A0A224XNR4 E2ANM6 A0A151IZ20 A0A195AU89 A0A158P1H7 A0A195CU90 F4WZ82 A0A195F6N1 A0A151X782 A0A182TX63 A0A182KNV0 A0A182X5F2 Q7Q5V6 A0A182I193 A0A182VFR2 A0A1B0DQ62 A0A2J7PQ66 A0A182YQL6 E2BNY8 A0A084WL39 A0A1Q3FR24 B0WZL3 A0A1J1J072 W5JD69 A0A182QI75 A0A0K8T373 A0A182GNH2 T1DCS2 A0A182GRX0 E0VHV4 A0A1S4FWD6 A0A2J7PQ75 A0A182FHZ3 Q16M66 A0A2M4BL91 A0A182PAY8 A0A336KPL9 A0A182NK04 T1HRS8 A0A0L7R9Y1 U5EQI6 A0A0C9Q806 A0A182WLN6 A0A154PG02 W8BH76 A0A310SJL2 A0A034V0W4 V9IDJ2 A0A2A3ECP6 A0A0A1XR59 A0A0N0U4Y2 A0A2S2PE15 J9JKE1 A0A088AD75 A0A2H8TVJ7 A0A2S2QW30 A0A0L0CR85 B4MPQ1 A0A1I8MQU4 A0A1I8MQT4 A0A0J9R7Q2 A0A0J9TVW8 A0A1W4VZE3 A0A1W4WB39 B4P278 B3N9A2 A0A0Q5WCH7 A0A0R1DTD0 B4J829 E1NZE3 A0A1I8PZ34 A0A1I8PZ04 Q7K2X5 B3MGD0 D0Z756 Q290X6 A0A3B0JVR9 A0A3B0J246 B4KQ79 A0A0Q9X8N7 A0A0Q9WFH4 B4LJK8 A0A0M4EBR5 B4GB29
A0A1B6LR65 A0A1B6GGD2 A0A1B6GL60 A0A1B6DK70 U4U2K4 N6UFH3 A0A139WCJ5 A0A1Y1LFZ3 A0A026X1V6 A0A224XNR4 E2ANM6 A0A151IZ20 A0A195AU89 A0A158P1H7 A0A195CU90 F4WZ82 A0A195F6N1 A0A151X782 A0A182TX63 A0A182KNV0 A0A182X5F2 Q7Q5V6 A0A182I193 A0A182VFR2 A0A1B0DQ62 A0A2J7PQ66 A0A182YQL6 E2BNY8 A0A084WL39 A0A1Q3FR24 B0WZL3 A0A1J1J072 W5JD69 A0A182QI75 A0A0K8T373 A0A182GNH2 T1DCS2 A0A182GRX0 E0VHV4 A0A1S4FWD6 A0A2J7PQ75 A0A182FHZ3 Q16M66 A0A2M4BL91 A0A182PAY8 A0A336KPL9 A0A182NK04 T1HRS8 A0A0L7R9Y1 U5EQI6 A0A0C9Q806 A0A182WLN6 A0A154PG02 W8BH76 A0A310SJL2 A0A034V0W4 V9IDJ2 A0A2A3ECP6 A0A0A1XR59 A0A0N0U4Y2 A0A2S2PE15 J9JKE1 A0A088AD75 A0A2H8TVJ7 A0A2S2QW30 A0A0L0CR85 B4MPQ1 A0A1I8MQU4 A0A1I8MQT4 A0A0J9R7Q2 A0A0J9TVW8 A0A1W4VZE3 A0A1W4WB39 B4P278 B3N9A2 A0A0Q5WCH7 A0A0R1DTD0 B4J829 E1NZE3 A0A1I8PZ34 A0A1I8PZ04 Q7K2X5 B3MGD0 D0Z756 Q290X6 A0A3B0JVR9 A0A3B0J246 B4KQ79 A0A0Q9X8N7 A0A0Q9WFH4 B4LJK8 A0A0M4EBR5 B4GB29
Pubmed
22118469
19121390
26354079
26227816
23537049
18362917
+ More
19820115 28004739 24508170 30249741 20798317 21347285 21719571 20966253 12364791 14747013 17210077 25244985 24438588 20920257 23761445 26483478 24330624 20566863 17510324 24495485 25348373 25830018 26108605 17994087 25315136 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18057021
19820115 28004739 24508170 30249741 20798317 21347285 21719571 20966253 12364791 14747013 17210077 25244985 24438588 20920257 23761445 26483478 24330624 20566863 17510324 24495485 25348373 25830018 26108605 17994087 25315136 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18057021
EMBL
NWSH01000113
PCG79424.1
AGBW02012900
OWR44241.1
BABH01008578
BABH01008579
+ More
BABH01008580 BABH01008581 KQ459606 KPI91647.1 JTDY01000040 KOB79231.1 KQ461198 KPJ06240.1 GEBQ01013840 GEBQ01004518 JAT26137.1 JAT35459.1 GECZ01008265 JAS61504.1 GECZ01006592 JAS63177.1 GEDC01011205 GEDC01003484 GEDC01003003 JAS26093.1 JAS33814.1 JAS34295.1 KB631301 KB631558 ERL84230.1 ERL84383.1 APGK01023871 KB740523 ENN80480.1 KQ971366 KYB25652.1 GEZM01056961 GEZM01056960 JAV72544.1 KK107024 QOIP01000004 EZA62290.1 RLU24053.1 GFTR01006765 JAW09661.1 GL441256 EFN64972.1 KQ980735 KYN13747.1 KQ976741 KYM75590.1 ADTU01006633 ADTU01006634 KQ977305 KYN03704.1 GL888470 EGI60471.1 KQ981756 KYN36118.1 KQ982450 KYQ56232.1 AAAB01008960 EAA11472.3 APCN01000045 AJVK01008455 AJVK01008456 NEVH01022640 PNF18482.1 GL449511 EFN82580.1 ATLV01024179 KE525350 KFB50933.1 GFDL01005041 JAV30004.1 DS232210 EDS37527.1 CVRI01000066 CRL05923.1 ADMH02001577 ETN62006.1 AXCN02000495 GBRD01006206 JAG59615.1 JXUM01000501 JXUM01000502 JXUM01000503 JXUM01000504 JXUM01000505 JXUM01000506 GALA01001727 JAA93125.1 JXUM01083457 JXUM01083458 JXUM01083459 JXUM01083460 JXUM01083461 KQ563378 KXJ73948.1 DS235172 EEB12960.1 PNF18483.1 CH477878 EAT35420.1 GGFJ01004580 MBW53721.1 UFQS01000669 UFQT01000669 SSX05978.1 SSX26335.1 ACPB03008601 ACPB03008602 ACPB03008603 KQ414620 KOC67675.1 GANO01004268 JAB55603.1 GBYB01010293 GBYB01010294 JAG80060.1 JAG80061.1 KQ434889 KZC10364.1 GAMC01017563 GAMC01017562 GAMC01017561 JAB88994.1 KQ764772 OAD54386.1 GAKP01022044 GAKP01022041 JAC36911.1 JR040449 JR040450 AEY59139.1 AEY59140.1 KZ288285 PBC29498.1 GBXI01002265 GBXI01000518 JAD12027.1 JAD13774.1 KQ435811 KOX72921.1 GGMR01015072 MBY27691.1 ABLF02028970 ABLF02028971 ABLF02028973 GFXV01006490 MBW18295.1 GGMS01012766 MBY81969.1 JRES01000023 KNC34848.1 CH963849 EDW74090.1 CM002911 KMY92166.1 KMY92167.1 KMY92165.1 CM000157 EDW89279.1 CH954177 EDV59589.1 KQS70900.1 KRJ98345.1 CH916367 EDW02259.1 BT125692 ADO16258.1 AE013599 AY060601 AAF59148.2 AAL28149.1 CH902619 EDV37833.2 BT100303 ACZ48702.1 AFH07953.1 CM000071 EAL25236.3 OUUW01000001 SPP75168.1 SPP75167.1 CH933808 EDW09207.2 KRG04525.1 KRG04526.1 CH940648 KRF80079.1 EDW61576.2 KRF80080.1 CP012524 ALC41027.1 CH479181 EDW32131.1
BABH01008580 BABH01008581 KQ459606 KPI91647.1 JTDY01000040 KOB79231.1 KQ461198 KPJ06240.1 GEBQ01013840 GEBQ01004518 JAT26137.1 JAT35459.1 GECZ01008265 JAS61504.1 GECZ01006592 JAS63177.1 GEDC01011205 GEDC01003484 GEDC01003003 JAS26093.1 JAS33814.1 JAS34295.1 KB631301 KB631558 ERL84230.1 ERL84383.1 APGK01023871 KB740523 ENN80480.1 KQ971366 KYB25652.1 GEZM01056961 GEZM01056960 JAV72544.1 KK107024 QOIP01000004 EZA62290.1 RLU24053.1 GFTR01006765 JAW09661.1 GL441256 EFN64972.1 KQ980735 KYN13747.1 KQ976741 KYM75590.1 ADTU01006633 ADTU01006634 KQ977305 KYN03704.1 GL888470 EGI60471.1 KQ981756 KYN36118.1 KQ982450 KYQ56232.1 AAAB01008960 EAA11472.3 APCN01000045 AJVK01008455 AJVK01008456 NEVH01022640 PNF18482.1 GL449511 EFN82580.1 ATLV01024179 KE525350 KFB50933.1 GFDL01005041 JAV30004.1 DS232210 EDS37527.1 CVRI01000066 CRL05923.1 ADMH02001577 ETN62006.1 AXCN02000495 GBRD01006206 JAG59615.1 JXUM01000501 JXUM01000502 JXUM01000503 JXUM01000504 JXUM01000505 JXUM01000506 GALA01001727 JAA93125.1 JXUM01083457 JXUM01083458 JXUM01083459 JXUM01083460 JXUM01083461 KQ563378 KXJ73948.1 DS235172 EEB12960.1 PNF18483.1 CH477878 EAT35420.1 GGFJ01004580 MBW53721.1 UFQS01000669 UFQT01000669 SSX05978.1 SSX26335.1 ACPB03008601 ACPB03008602 ACPB03008603 KQ414620 KOC67675.1 GANO01004268 JAB55603.1 GBYB01010293 GBYB01010294 JAG80060.1 JAG80061.1 KQ434889 KZC10364.1 GAMC01017563 GAMC01017562 GAMC01017561 JAB88994.1 KQ764772 OAD54386.1 GAKP01022044 GAKP01022041 JAC36911.1 JR040449 JR040450 AEY59139.1 AEY59140.1 KZ288285 PBC29498.1 GBXI01002265 GBXI01000518 JAD12027.1 JAD13774.1 KQ435811 KOX72921.1 GGMR01015072 MBY27691.1 ABLF02028970 ABLF02028971 ABLF02028973 GFXV01006490 MBW18295.1 GGMS01012766 MBY81969.1 JRES01000023 KNC34848.1 CH963849 EDW74090.1 CM002911 KMY92166.1 KMY92167.1 KMY92165.1 CM000157 EDW89279.1 CH954177 EDV59589.1 KQS70900.1 KRJ98345.1 CH916367 EDW02259.1 BT125692 ADO16258.1 AE013599 AY060601 AAF59148.2 AAL28149.1 CH902619 EDV37833.2 BT100303 ACZ48702.1 AFH07953.1 CM000071 EAL25236.3 OUUW01000001 SPP75168.1 SPP75167.1 CH933808 EDW09207.2 KRG04525.1 KRG04526.1 CH940648 KRF80079.1 EDW61576.2 KRF80080.1 CP012524 ALC41027.1 CH479181 EDW32131.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053268
UP000037510
UP000053240
+ More
UP000030742 UP000019118 UP000007266 UP000053097 UP000279307 UP000000311 UP000078492 UP000078540 UP000005205 UP000078542 UP000007755 UP000078541 UP000075809 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075903 UP000092462 UP000235965 UP000076408 UP000008237 UP000030765 UP000002320 UP000183832 UP000000673 UP000075886 UP000069940 UP000249989 UP000009046 UP000069272 UP000008820 UP000075885 UP000075884 UP000015103 UP000053825 UP000075920 UP000076502 UP000242457 UP000053105 UP000007819 UP000005203 UP000037069 UP000007798 UP000095301 UP000192221 UP000002282 UP000008711 UP000001070 UP000095300 UP000000803 UP000007801 UP000001819 UP000268350 UP000009192 UP000008792 UP000092553 UP000008744
UP000030742 UP000019118 UP000007266 UP000053097 UP000279307 UP000000311 UP000078492 UP000078540 UP000005205 UP000078542 UP000007755 UP000078541 UP000075809 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075903 UP000092462 UP000235965 UP000076408 UP000008237 UP000030765 UP000002320 UP000183832 UP000000673 UP000075886 UP000069940 UP000249989 UP000009046 UP000069272 UP000008820 UP000075885 UP000075884 UP000015103 UP000053825 UP000075920 UP000076502 UP000242457 UP000053105 UP000007819 UP000005203 UP000037069 UP000007798 UP000095301 UP000192221 UP000002282 UP000008711 UP000001070 UP000095300 UP000000803 UP000007801 UP000001819 UP000268350 UP000009192 UP000008792 UP000092553 UP000008744
Interpro
SUPFAM
SSF57667
SSF57667
Gene 3D
ProteinModelPortal
A0A2A4K690
A0A212ES11
H9JAQ6
A0A194PEF0
A0A0L7LVM3
A0A194QMQ2
+ More
A0A1B6LR65 A0A1B6GGD2 A0A1B6GL60 A0A1B6DK70 U4U2K4 N6UFH3 A0A139WCJ5 A0A1Y1LFZ3 A0A026X1V6 A0A224XNR4 E2ANM6 A0A151IZ20 A0A195AU89 A0A158P1H7 A0A195CU90 F4WZ82 A0A195F6N1 A0A151X782 A0A182TX63 A0A182KNV0 A0A182X5F2 Q7Q5V6 A0A182I193 A0A182VFR2 A0A1B0DQ62 A0A2J7PQ66 A0A182YQL6 E2BNY8 A0A084WL39 A0A1Q3FR24 B0WZL3 A0A1J1J072 W5JD69 A0A182QI75 A0A0K8T373 A0A182GNH2 T1DCS2 A0A182GRX0 E0VHV4 A0A1S4FWD6 A0A2J7PQ75 A0A182FHZ3 Q16M66 A0A2M4BL91 A0A182PAY8 A0A336KPL9 A0A182NK04 T1HRS8 A0A0L7R9Y1 U5EQI6 A0A0C9Q806 A0A182WLN6 A0A154PG02 W8BH76 A0A310SJL2 A0A034V0W4 V9IDJ2 A0A2A3ECP6 A0A0A1XR59 A0A0N0U4Y2 A0A2S2PE15 J9JKE1 A0A088AD75 A0A2H8TVJ7 A0A2S2QW30 A0A0L0CR85 B4MPQ1 A0A1I8MQU4 A0A1I8MQT4 A0A0J9R7Q2 A0A0J9TVW8 A0A1W4VZE3 A0A1W4WB39 B4P278 B3N9A2 A0A0Q5WCH7 A0A0R1DTD0 B4J829 E1NZE3 A0A1I8PZ34 A0A1I8PZ04 Q7K2X5 B3MGD0 D0Z756 Q290X6 A0A3B0JVR9 A0A3B0J246 B4KQ79 A0A0Q9X8N7 A0A0Q9WFH4 B4LJK8 A0A0M4EBR5 B4GB29
A0A1B6LR65 A0A1B6GGD2 A0A1B6GL60 A0A1B6DK70 U4U2K4 N6UFH3 A0A139WCJ5 A0A1Y1LFZ3 A0A026X1V6 A0A224XNR4 E2ANM6 A0A151IZ20 A0A195AU89 A0A158P1H7 A0A195CU90 F4WZ82 A0A195F6N1 A0A151X782 A0A182TX63 A0A182KNV0 A0A182X5F2 Q7Q5V6 A0A182I193 A0A182VFR2 A0A1B0DQ62 A0A2J7PQ66 A0A182YQL6 E2BNY8 A0A084WL39 A0A1Q3FR24 B0WZL3 A0A1J1J072 W5JD69 A0A182QI75 A0A0K8T373 A0A182GNH2 T1DCS2 A0A182GRX0 E0VHV4 A0A1S4FWD6 A0A2J7PQ75 A0A182FHZ3 Q16M66 A0A2M4BL91 A0A182PAY8 A0A336KPL9 A0A182NK04 T1HRS8 A0A0L7R9Y1 U5EQI6 A0A0C9Q806 A0A182WLN6 A0A154PG02 W8BH76 A0A310SJL2 A0A034V0W4 V9IDJ2 A0A2A3ECP6 A0A0A1XR59 A0A0N0U4Y2 A0A2S2PE15 J9JKE1 A0A088AD75 A0A2H8TVJ7 A0A2S2QW30 A0A0L0CR85 B4MPQ1 A0A1I8MQU4 A0A1I8MQT4 A0A0J9R7Q2 A0A0J9TVW8 A0A1W4VZE3 A0A1W4WB39 B4P278 B3N9A2 A0A0Q5WCH7 A0A0R1DTD0 B4J829 E1NZE3 A0A1I8PZ34 A0A1I8PZ04 Q7K2X5 B3MGD0 D0Z756 Q290X6 A0A3B0JVR9 A0A3B0J246 B4KQ79 A0A0Q9X8N7 A0A0Q9WFH4 B4LJK8 A0A0M4EBR5 B4GB29
PDB
3KJ4
E-value=6.24273e-19,
Score=232
Ontologies
GO
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.997628
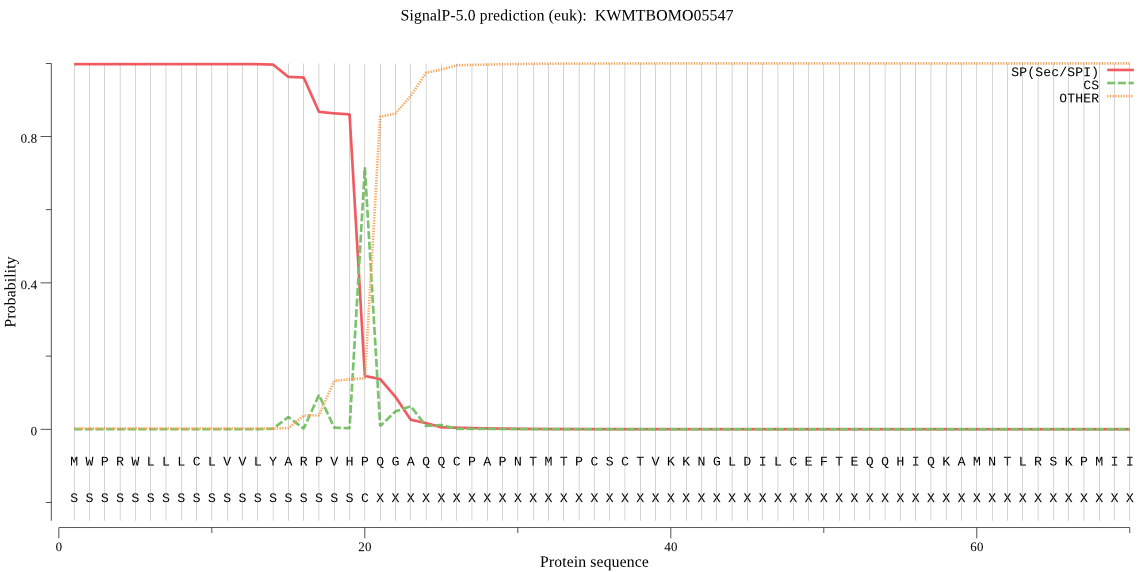
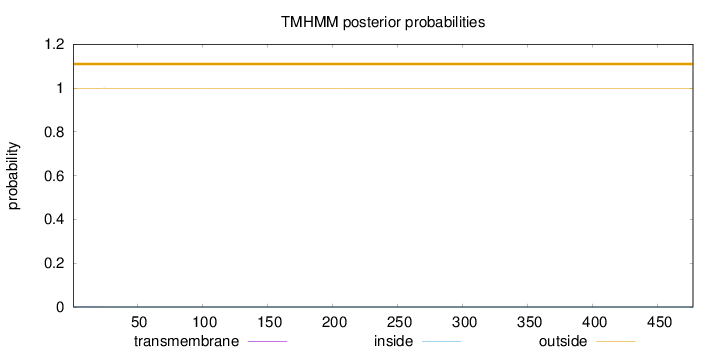
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01605
Exp number, first 60 AAs:
0.0142
Total prob of N-in:
0.00134
outside
1 - 477
Population Genetic Test Statistics
Pi
24.678855
Theta
182.500888
Tajima's D
0.459254
CLR
1.420809
CSRT
0.501824908754562
Interpretation
Uncertain
