Gene
KWMTBOMO05546
Pre Gene Modal
BGIBMGA006740
Annotation
PREDICTED:_laccase-4-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.122
Sequence
CDS
ATGGAGCCGCCGAAAACCGCGATGGAAGTGCACGGTGTGCCAGTGCAGAAACCAAATTACATGCTCATGTCCTTACAAAGGATATTAGTGCTTGTTATTATATTCGCATGCGTTCTAATTGTAGTGCGCTACACACCGATGCCTGAAGAATACTTTGAAAACTGCGACAGAGAATGTCATGAATTAGACTGGCCAATGATTTGTCGTGTGAAACTAGTAATCGAAGTCTACAAAACTCTTAGCAAATCTTGTGGCAATTGTACTGATAAAGATGGAACAGGAGAGTGTCCGTCCACGTGTATATCCGCAGACGGACGGGAGCGAGGTGTTCTCACAGCGAACCGTGCTTTACCAGCACCAACGCTTCACGTCTGTCACAACGATATCCTCGTAGTCGATGTAGTGCATAGAGCTCCGGCTCATGCGTTATCTATCCATTGGCGCGGTCAACCACAAAAGGAAACGCCCTTTATGGACGGAGCCCCAATGTTGACACAATGCCCGCAACCTGCATATACTACATTTCAATATAAATTCCGAGCATCTGCAGTTGGAACACATATGTACCACGCTCACTCCGCAGCGGACGCTGCTGATGGTCTTGCAGGCGCCCTTGTGGTCCGACAATCTCCTCGTCAAGACCCATTGAGGAAACTGTACGACACTGATGCTTCGGAGCATACAATATATGTTTCCGAATGGGGTCATTCAATGGGACCCTTGGCGGGTATGATGTCGAATGTTCCTAAAGCTGAATCGCTTCTCATAAACGGGAAAGGAAAGACCGTTGAAAATATAGATGCACCGTTAACTAAGTTTTCCGTTGAATATGGAAAGCGGTATAGATTTAGATTAGCATACGGTGGAGGATCGAAGAGTTGTCCTATTCAATTTTCTATTGAAAAACACGTTCTCACTCTGGTCGCTTTAGATGGAAATGGAATAGAACCGGAATATGTTAATTCTATTGAATTAGGTCGAGGGGAGCGTGTTGATTTTATACTAGAAGCCAAAAGAGCTCCGGGCGTATACAAAATGTCCGTTGTGGCGCATCCGGATTGTCAAGATAATTTAAAAGGTGTAGCTGAGTTGGTTTATACAAATAAAGATAAAGCAGTGTTAATTAAAGACATTGACGAGTTAAATAAAGTTTATCGTAAGTTTAGCACAGTGATTAGTGTGCAGTGTGAGGACGAAAGCGTCTTGTGTTTGACAGAGGCTCGTGGCTATGAGCAGATGCCCTCTGAACTGACAAAGACTCCCGACCGGACTCTCTATGTGCCTTTTAATTATTCCACAAGAAGAATATCAGCGAAGCGAGTCGAGAGTTGGGGTCAATCTGACGGTCATCGTTTCACATATCCGGCATCTCCACTGCTCACTCAAGGAGGCGACGTGGGACCCAACACTTTGTGTCCTGAAGGGCCCGGTGAGGGCGACGAATGCGTCCATGTTAAAAATATTCCATTGTATTCTACCGTGGAAATTGTTATGTTTGATCAAGGTGGCGAGTCGGACCACATTTTCCATCTGCACGGATACGGTTTCTACGTTACCGGAGTGAGGGAGTTTAACAGGAGTCTGAGTAAGGAAACAGTCATCAAAATGAACGAAGCGAACACGCTATTTATAAACAAGAATCTCATCAGACCCGTATTAAAAGACACTATCGTGATACCTAAATTCGGCGTAGTCGCGTTGCGGTTTAAGGCGGATAATCCGGGGTACTGGATGATGAGGGACGAAAGGTCGACCCACTGGACAAGAGGACTCGATTTCATATTAAAAGTGGGCGAACAAAGCGACTTAGTGCAGGCGCCTCCTGATTTTCCGAAATGTGGTTCATATGTCGGCCCCGAGTATTTTCTAATTTAA
Protein
MEPPKTAMEVHGVPVQKPNYMLMSLQRILVLVIIFACVLIVVRYTPMPEEYFENCDRECHELDWPMICRVKLVIEVYKTLSKSCGNCTDKDGTGECPSTCISADGRERGVLTANRALPAPTLHVCHNDILVVDVVHRAPAHALSIHWRGQPQKETPFMDGAPMLTQCPQPAYTTFQYKFRASAVGTHMYHAHSAADAADGLAGALVVRQSPRQDPLRKLYDTDASEHTIYVSEWGHSMGPLAGMMSNVPKAESLLINGKGKTVENIDAPLTKFSVEYGKRYRFRLAYGGGSKSCPIQFSIEKHVLTLVALDGNGIEPEYVNSIELGRGERVDFILEAKRAPGVYKMSVVAHPDCQDNLKGVAELVYTNKDKAVLIKDIDELNKVYRKFSTVISVQCEDESVLCLTEARGYEQMPSELTKTPDRTLYVPFNYSTRRISAKRVESWGQSDGHRFTYPASPLLTQGGDVGPNTLCPEGPGEGDECVHVKNIPLYSTVEIVMFDQGGESDHIFHLHGYGFYVTGVREFNRSLSKETVIKMNEANTLFINKNLIRPVLKDTIVIPKFGVVALRFKADNPGYWMMRDERSTHWTRGLDFILKVGEQSDLVQAPPDFPKCGSYVGPEYFLI
Summary
Similarity
Belongs to the multicopper oxidase family.
Uniprot
A0A2A4KA06
A0A194PF48
A0A3S2LSV4
A0A212F814
A0A194QRS4
H9JB45
+ More
A0A2H1WX52 A0A0L7L9Z9 A0A195F694 A0A195CSL7 A0A151X7F6 A0A151IYW8 A0A158P1H9 E2BNY5 A0A3L8DUC3 A0A0M9A2N5 A0A0L7RA16 A0A154PEY5 E2ANM3 A0A310SQT5 A0A067QPY4 A0A0H3YIT1 A0A195ATN5 A0A1S6J0Y4 A0A1Y1LIX3 A0A0T6BI01 A0A0A9WZS0 A0A146L2B6 D2CG53 A0A2Z5WQ00 A0A1B6FDH2 U4U336 A0A026X4G0 A0A1B6E1C2 A0A024B7C8 A0A146M7B7 E0VD01 N6UP17 U4U109 A0A0C9PX96 A0A2J7QYY4 A0A1B6JHU7 J9JQ50 A0A2S2NBT2 A0A2S2QQW2 A0A088AD72 E9I913 T1JM48 A0A2P8XLJ7 A0A067QYK5 A0A1B6FJ43 A0A0P4YD86 A0A0P4XYI2 A0A1A9WAB1 A0A067QPY0 A0A0P6AJZ3 A0A3S2N8B0 A0A182M2S5 E9GEP8 A0A2A4K7B1 A0A2W1BMF7 A0A0P5SEH3 A0A0F7LJ96 A0A2J7QYW3 A0A1B6IUJ5 A0A0P5VN40 A0A0P5TNK5 D5MRE2 A0A0P5LTX1 A0A0P6CZG5 B4F7L5 A0A0P5TC11 A0A0P6JN49 A0A0P6AWC6 A0A0P5TVL7 A0A0N8CHB3 A0A1B0C0D4 A0A0P5SWM3 A0A1A9Y962 A0A1A9ULU8 A0A0P5E4P7 A0A0P6ES77 A0A291KZC6 Q8I8Y1 A0A1A9ZUT2 A0A164MPJ0 A0A0N7ZKA7 A0A182F3R6 A0A0P5CDY8 A0A0P4Y4Z7 A0A2J7PQ10 A0A0P4Y7D9 A0A0N8AHB9 A0A2J7PQ17 A0A0P4ZCE1 A0A0P4XGH4 A0A0P4XLY4 A0A0P5E6R2 A0A0P5ZP30
A0A2H1WX52 A0A0L7L9Z9 A0A195F694 A0A195CSL7 A0A151X7F6 A0A151IYW8 A0A158P1H9 E2BNY5 A0A3L8DUC3 A0A0M9A2N5 A0A0L7RA16 A0A154PEY5 E2ANM3 A0A310SQT5 A0A067QPY4 A0A0H3YIT1 A0A195ATN5 A0A1S6J0Y4 A0A1Y1LIX3 A0A0T6BI01 A0A0A9WZS0 A0A146L2B6 D2CG53 A0A2Z5WQ00 A0A1B6FDH2 U4U336 A0A026X4G0 A0A1B6E1C2 A0A024B7C8 A0A146M7B7 E0VD01 N6UP17 U4U109 A0A0C9PX96 A0A2J7QYY4 A0A1B6JHU7 J9JQ50 A0A2S2NBT2 A0A2S2QQW2 A0A088AD72 E9I913 T1JM48 A0A2P8XLJ7 A0A067QYK5 A0A1B6FJ43 A0A0P4YD86 A0A0P4XYI2 A0A1A9WAB1 A0A067QPY0 A0A0P6AJZ3 A0A3S2N8B0 A0A182M2S5 E9GEP8 A0A2A4K7B1 A0A2W1BMF7 A0A0P5SEH3 A0A0F7LJ96 A0A2J7QYW3 A0A1B6IUJ5 A0A0P5VN40 A0A0P5TNK5 D5MRE2 A0A0P5LTX1 A0A0P6CZG5 B4F7L5 A0A0P5TC11 A0A0P6JN49 A0A0P6AWC6 A0A0P5TVL7 A0A0N8CHB3 A0A1B0C0D4 A0A0P5SWM3 A0A1A9Y962 A0A1A9ULU8 A0A0P5E4P7 A0A0P6ES77 A0A291KZC6 Q8I8Y1 A0A1A9ZUT2 A0A164MPJ0 A0A0N7ZKA7 A0A182F3R6 A0A0P5CDY8 A0A0P4Y4Z7 A0A2J7PQ10 A0A0P4Y7D9 A0A0N8AHB9 A0A2J7PQ17 A0A0P4ZCE1 A0A0P4XGH4 A0A0P4XLY4 A0A0P5E6R2 A0A0P5ZP30
Pubmed
EMBL
NWSH01000001
PCG81125.1
KQ459606
KPI91648.1
RSAL01000015
RVE53119.1
+ More
AGBW02009807 OWR49884.1 KQ461198 KPJ06241.1 BABH01008575 ODYU01011676 SOQ57572.1 JTDY01002063 KOB72200.1 KQ981756 KYN36115.1 KQ977305 KYN03708.1 KQ982450 KYQ56228.1 KQ980735 KYN13750.1 ADTU01006647 ADTU01006648 GL449511 EFN82577.1 QOIP01000004 RLU24060.1 KQ435755 KOX76045.1 KQ414620 KOC67679.1 KQ434889 KZC10367.1 GL441256 EFN64969.1 KQ759903 OAD62006.1 KK853076 KDR11754.1 KR086791 AKN21384.1 KQ976741 KYM75588.1 KU764432 AQS60678.1 GEZM01056202 JAV72788.1 LJIG01000043 KRT86897.1 GBHO01030380 GBHO01030378 JAG13224.1 JAG13226.1 GDHC01017173 JAQ01456.1 KQ971366 EFA12828.2 LC202058 BBC20926.1 GECZ01021538 JAS48231.1 KB631557 ERL84380.1 KK107024 EZA62294.1 GEDC01005594 JAS31704.1 KJ500311 AHZ13215.1 GDHC01003310 JAQ15319.1 DS235068 EEB11257.1 APGK01023870 KB740523 ENN80477.1 KB631857 ERL86762.1 GBYB01006133 JAG75900.1 NEVH01009082 PNF33797.1 GECU01008876 GECU01008309 JAS98830.1 JAS99397.1 ABLF02034333 GGMR01002055 MBY14674.1 GGMS01010717 MBY79920.1 GL761667 EFZ22956.1 JH431603 PYGN01001776 PSN32868.1 KK852824 KDR15437.1 GECZ01019605 JAS50164.1 GDIP01238862 JAI84539.1 GDIP01238861 JAI84540.1 KDR11749.1 GDIP01033644 JAM70071.1 RSAL01000212 RVE44181.1 AXCM01012146 GL732541 EFX81873.1 NWSH01000053 PCG80165.1 KZ150026 PZC74805.1 GDIP01140785 JAL62929.1 KP318028 AKH61981.1 PNF33770.1 GECU01017108 JAS90598.1 GDIP01097790 JAM05925.1 GDIP01141440 JAL62274.1 AB514130 BAJ06132.1 GDIQ01165221 JAK86504.1 GDIQ01087496 JAN07241.1 BK006377 DAA06286.1 GDIP01210175 GDIP01175564 GDIP01147144 GDIP01146474 GDIP01134236 GDIP01131744 GDIP01130384 GDIP01051957 JAL69478.1 GDIQ01013899 JAN80838.1 GDIP01025604 JAM78111.1 GDIP01126090 JAL77624.1 GDIP01241175 GDIP01232184 GDIP01230924 GDIP01222080 GDIP01220930 GDIP01167326 GDIP01165924 GDIP01147145 GDIP01138874 GDIP01134235 GDIP01131743 GDIP01120831 GDIP01074054 GDIP01064047 GDIP01044646 JAL71971.1 JXJN01023535 GDIP01241174 GDIP01232183 GDIP01230923 GDIP01222079 GDIP01220931 GDIP01185570 GDIP01134237 GDIP01124992 GDIP01064046 LRGB01000915 JAL69477.1 KZS15189.1 GDIP01165125 JAJ58277.1 GDIQ01071949 JAN22788.1 KY073340 ATI08981.1 AY135185 AAN17506.1 LRGB01002985 KZS05241.1 GDIP01237081 JAI86320.1 GDIP01176778 JAJ46624.1 GDIP01232520 GDIP01232519 JAI90881.1 NEVH01022640 PNF18422.1 GDIP01232518 JAI90883.1 GDIP01147385 JAJ76017.1 PNF18421.1 GDIP01227818 JAI95583.1 GDIP01241462 GDIP01241461 JAI81940.1 GDIP01239973 GDIP01239972 GDIP01044793 JAI83429.1 GDIP01147386 JAJ76016.1 GDIP01041092 JAM62623.1
AGBW02009807 OWR49884.1 KQ461198 KPJ06241.1 BABH01008575 ODYU01011676 SOQ57572.1 JTDY01002063 KOB72200.1 KQ981756 KYN36115.1 KQ977305 KYN03708.1 KQ982450 KYQ56228.1 KQ980735 KYN13750.1 ADTU01006647 ADTU01006648 GL449511 EFN82577.1 QOIP01000004 RLU24060.1 KQ435755 KOX76045.1 KQ414620 KOC67679.1 KQ434889 KZC10367.1 GL441256 EFN64969.1 KQ759903 OAD62006.1 KK853076 KDR11754.1 KR086791 AKN21384.1 KQ976741 KYM75588.1 KU764432 AQS60678.1 GEZM01056202 JAV72788.1 LJIG01000043 KRT86897.1 GBHO01030380 GBHO01030378 JAG13224.1 JAG13226.1 GDHC01017173 JAQ01456.1 KQ971366 EFA12828.2 LC202058 BBC20926.1 GECZ01021538 JAS48231.1 KB631557 ERL84380.1 KK107024 EZA62294.1 GEDC01005594 JAS31704.1 KJ500311 AHZ13215.1 GDHC01003310 JAQ15319.1 DS235068 EEB11257.1 APGK01023870 KB740523 ENN80477.1 KB631857 ERL86762.1 GBYB01006133 JAG75900.1 NEVH01009082 PNF33797.1 GECU01008876 GECU01008309 JAS98830.1 JAS99397.1 ABLF02034333 GGMR01002055 MBY14674.1 GGMS01010717 MBY79920.1 GL761667 EFZ22956.1 JH431603 PYGN01001776 PSN32868.1 KK852824 KDR15437.1 GECZ01019605 JAS50164.1 GDIP01238862 JAI84539.1 GDIP01238861 JAI84540.1 KDR11749.1 GDIP01033644 JAM70071.1 RSAL01000212 RVE44181.1 AXCM01012146 GL732541 EFX81873.1 NWSH01000053 PCG80165.1 KZ150026 PZC74805.1 GDIP01140785 JAL62929.1 KP318028 AKH61981.1 PNF33770.1 GECU01017108 JAS90598.1 GDIP01097790 JAM05925.1 GDIP01141440 JAL62274.1 AB514130 BAJ06132.1 GDIQ01165221 JAK86504.1 GDIQ01087496 JAN07241.1 BK006377 DAA06286.1 GDIP01210175 GDIP01175564 GDIP01147144 GDIP01146474 GDIP01134236 GDIP01131744 GDIP01130384 GDIP01051957 JAL69478.1 GDIQ01013899 JAN80838.1 GDIP01025604 JAM78111.1 GDIP01126090 JAL77624.1 GDIP01241175 GDIP01232184 GDIP01230924 GDIP01222080 GDIP01220930 GDIP01167326 GDIP01165924 GDIP01147145 GDIP01138874 GDIP01134235 GDIP01131743 GDIP01120831 GDIP01074054 GDIP01064047 GDIP01044646 JAL71971.1 JXJN01023535 GDIP01241174 GDIP01232183 GDIP01230923 GDIP01222079 GDIP01220931 GDIP01185570 GDIP01134237 GDIP01124992 GDIP01064046 LRGB01000915 JAL69477.1 KZS15189.1 GDIP01165125 JAJ58277.1 GDIQ01071949 JAN22788.1 KY073340 ATI08981.1 AY135185 AAN17506.1 LRGB01002985 KZS05241.1 GDIP01237081 JAI86320.1 GDIP01176778 JAJ46624.1 GDIP01232520 GDIP01232519 JAI90881.1 NEVH01022640 PNF18422.1 GDIP01232518 JAI90883.1 GDIP01147385 JAJ76017.1 PNF18421.1 GDIP01227818 JAI95583.1 GDIP01241462 GDIP01241461 JAI81940.1 GDIP01239973 GDIP01239972 GDIP01044793 JAI83429.1 GDIP01147386 JAJ76016.1 GDIP01041092 JAM62623.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
UP000005204
+ More
UP000037510 UP000078541 UP000078542 UP000075809 UP000078492 UP000005205 UP000008237 UP000279307 UP000053105 UP000053825 UP000076502 UP000000311 UP000027135 UP000078540 UP000007266 UP000030742 UP000053097 UP000009046 UP000019118 UP000235965 UP000007819 UP000005203 UP000245037 UP000091820 UP000075883 UP000000305 UP000092460 UP000076858 UP000092443 UP000078200 UP000092445 UP000069272
UP000037510 UP000078541 UP000078542 UP000075809 UP000078492 UP000005205 UP000008237 UP000279307 UP000053105 UP000053825 UP000076502 UP000000311 UP000027135 UP000078540 UP000007266 UP000030742 UP000053097 UP000009046 UP000019118 UP000235965 UP000007819 UP000005203 UP000245037 UP000091820 UP000075883 UP000000305 UP000092460 UP000076858 UP000092443 UP000078200 UP000092445 UP000069272
Interpro
SUPFAM
SSF49503
SSF49503
Gene 3D
ProteinModelPortal
A0A2A4KA06
A0A194PF48
A0A3S2LSV4
A0A212F814
A0A194QRS4
H9JB45
+ More
A0A2H1WX52 A0A0L7L9Z9 A0A195F694 A0A195CSL7 A0A151X7F6 A0A151IYW8 A0A158P1H9 E2BNY5 A0A3L8DUC3 A0A0M9A2N5 A0A0L7RA16 A0A154PEY5 E2ANM3 A0A310SQT5 A0A067QPY4 A0A0H3YIT1 A0A195ATN5 A0A1S6J0Y4 A0A1Y1LIX3 A0A0T6BI01 A0A0A9WZS0 A0A146L2B6 D2CG53 A0A2Z5WQ00 A0A1B6FDH2 U4U336 A0A026X4G0 A0A1B6E1C2 A0A024B7C8 A0A146M7B7 E0VD01 N6UP17 U4U109 A0A0C9PX96 A0A2J7QYY4 A0A1B6JHU7 J9JQ50 A0A2S2NBT2 A0A2S2QQW2 A0A088AD72 E9I913 T1JM48 A0A2P8XLJ7 A0A067QYK5 A0A1B6FJ43 A0A0P4YD86 A0A0P4XYI2 A0A1A9WAB1 A0A067QPY0 A0A0P6AJZ3 A0A3S2N8B0 A0A182M2S5 E9GEP8 A0A2A4K7B1 A0A2W1BMF7 A0A0P5SEH3 A0A0F7LJ96 A0A2J7QYW3 A0A1B6IUJ5 A0A0P5VN40 A0A0P5TNK5 D5MRE2 A0A0P5LTX1 A0A0P6CZG5 B4F7L5 A0A0P5TC11 A0A0P6JN49 A0A0P6AWC6 A0A0P5TVL7 A0A0N8CHB3 A0A1B0C0D4 A0A0P5SWM3 A0A1A9Y962 A0A1A9ULU8 A0A0P5E4P7 A0A0P6ES77 A0A291KZC6 Q8I8Y1 A0A1A9ZUT2 A0A164MPJ0 A0A0N7ZKA7 A0A182F3R6 A0A0P5CDY8 A0A0P4Y4Z7 A0A2J7PQ10 A0A0P4Y7D9 A0A0N8AHB9 A0A2J7PQ17 A0A0P4ZCE1 A0A0P4XGH4 A0A0P4XLY4 A0A0P5E6R2 A0A0P5ZP30
A0A2H1WX52 A0A0L7L9Z9 A0A195F694 A0A195CSL7 A0A151X7F6 A0A151IYW8 A0A158P1H9 E2BNY5 A0A3L8DUC3 A0A0M9A2N5 A0A0L7RA16 A0A154PEY5 E2ANM3 A0A310SQT5 A0A067QPY4 A0A0H3YIT1 A0A195ATN5 A0A1S6J0Y4 A0A1Y1LIX3 A0A0T6BI01 A0A0A9WZS0 A0A146L2B6 D2CG53 A0A2Z5WQ00 A0A1B6FDH2 U4U336 A0A026X4G0 A0A1B6E1C2 A0A024B7C8 A0A146M7B7 E0VD01 N6UP17 U4U109 A0A0C9PX96 A0A2J7QYY4 A0A1B6JHU7 J9JQ50 A0A2S2NBT2 A0A2S2QQW2 A0A088AD72 E9I913 T1JM48 A0A2P8XLJ7 A0A067QYK5 A0A1B6FJ43 A0A0P4YD86 A0A0P4XYI2 A0A1A9WAB1 A0A067QPY0 A0A0P6AJZ3 A0A3S2N8B0 A0A182M2S5 E9GEP8 A0A2A4K7B1 A0A2W1BMF7 A0A0P5SEH3 A0A0F7LJ96 A0A2J7QYW3 A0A1B6IUJ5 A0A0P5VN40 A0A0P5TNK5 D5MRE2 A0A0P5LTX1 A0A0P6CZG5 B4F7L5 A0A0P5TC11 A0A0P6JN49 A0A0P6AWC6 A0A0P5TVL7 A0A0N8CHB3 A0A1B0C0D4 A0A0P5SWM3 A0A1A9Y962 A0A1A9ULU8 A0A0P5E4P7 A0A0P6ES77 A0A291KZC6 Q8I8Y1 A0A1A9ZUT2 A0A164MPJ0 A0A0N7ZKA7 A0A182F3R6 A0A0P5CDY8 A0A0P4Y4Z7 A0A2J7PQ10 A0A0P4Y7D9 A0A0N8AHB9 A0A2J7PQ17 A0A0P4ZCE1 A0A0P4XGH4 A0A0P4XLY4 A0A0P5E6R2 A0A0P5ZP30
PDB
4JHV
E-value=1.89059e-34,
Score=367
Ontologies
GO
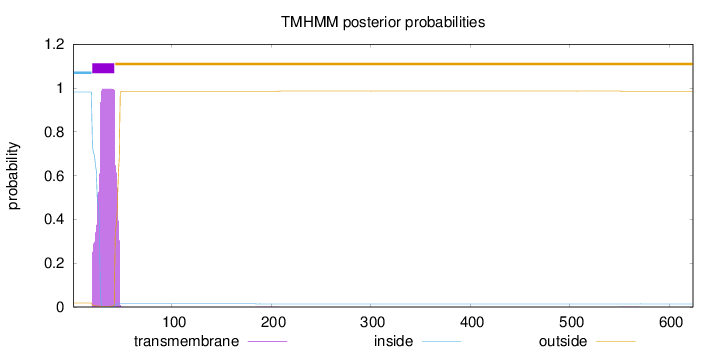
Topology
Length:
624
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.61174
Exp number, first 60 AAs:
20.59079
Total prob of N-in:
0.98195
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 624
Population Genetic Test Statistics
Pi
189.361905
Theta
165.188278
Tajima's D
0.088945
CLR
0.550375
CSRT
0.39718014099295
Interpretation
Uncertain
