Gene
KWMTBOMO05544
Pre Gene Modal
BGIBMGA006602
Annotation
PREDICTED:_poly(A)-specific_ribonuclease_PARN-like_domain-containing_protein_1_[Papilio_polytes]
Full name
Pre-piRNA 3'-exonuclease trimmer
Alternative Name
PARN-like domain-containing protein 1 homolog
Location in the cell
Nuclear Reliability : 2.231
Sequence
CDS
ATGGATATCACCAAAGAAAACTATTATGAAGAACTTGATAACATAGCAAAAAATATAAAAGACTCGTGTTTTATAAGTTTTGATGCAGAGTTTTCAGCTATACTTACCAAAGAATCGTTCAAATACAGCTTGTTCGACACAAATGAAGAAAGATACAATAAGTACAAAACACAAATAAGCACAATGAAAATGATGCAAGTAGGACTGACTATGTTTCAATATGATAGAGAATTAGATGCATACCTTGCAACTGGTTATACATTTCATTTGTGTCCACAACTAATTGGAACGATAAATCAGTCGCTCATATTTCAAGCCTCAACCCTCAAGTTTTTATGTAAACACAATTTCGATTTCAATAAGTTCATCTATGAAGGCCTTCCATATCTTAGCAAGAGTGATGAGGCATTGTTAAACAGATACAGAATAGAAAACAACCTGTTTGACTATGTCAGTGAGTCTCTTGAATTTGATGAAGAGAAGCAAATCAACCAATGTTCTTCAGAAGTTTCAAAATGGTTGTCATCGAGTATAGAGGACACAATGTACTTAGATATTGACAATGCTATTTGCCGGTACTTGCTGCACTTAGAGCTAAGACAACGTTATCCTGGTATCCTGACTACAGATAGCTTAGGTAATAGCAAGAAGGTAATTTAA
Protein
MDITKENYYEELDNIAKNIKDSCFISFDAEFSAILTKESFKYSLFDTNEERYNKYKTQISTMKMMQVGLTMFQYDRELDAYLATGYTFHLCPQLIGTINQSLIFQASTLKFLCKHNFDFNKFIYEGLPYLSKSDEALLNRYRIENNLFDYVSESLEFDEEKQINQCSSEVSKWLSSSIEDTMYLDIDNAICRYLLHLELRQRYPGILTTDSLGNSKKVI
Summary
Description
3'-5' exonuclease that specifically cleaves precursor piRNAs (pre-piRNAs) at their 3' ends (PubMed:26919431). Trims pre-piRNAs to their mature size, a process required for piRNAs maturation and stabilization, and subsequent pre-piRNAs 2'-O-methylation (PubMed:26919431). The piRNA metabolic process mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and govern the methylation and subsequent repression of transposons (PubMed:26919431).
Cofactor
Mg(2+)
Subunit
Interacts with Papi/Tdrkh; interaction takes place on the mitochondrial surface and recruits PNLDC1/trimmer to PIWI-bound pre-piRNAs.
Similarity
Belongs to the CAF1 family.
Keywords
Complete proteome
Exonuclease
Hydrolase
Magnesium
Membrane
Metal-binding
Mitochondrion
Mitochondrion outer membrane
Nuclease
Reference proteome
RNA-binding
RNA-mediated gene silencing
Transmembrane
Transmembrane helix
Feature
chain Pre-piRNA 3'-exonuclease trimmer
Uniprot
A0A1U6USN9
H9JAQ7
A0A3S2NZN8
A0A2A4KBR8
A0A194PG21
A0A194QL30
+ More
A0A212F817 A0A2H1WWY1 A0A0L7LAH1 A0A2J7R7Q0 A0A067RCE4 A0A2J7R7Q3 A0A2J7R7Q1 A0A2P8XTI4 A0A154PK67 A0A0J7L5J8 A0A3Q0J8T0 E2ANM1 A0A224XIL7 A0A0M9A428 A0A1Z5KY51 A0A2R5LL04 A0A2A3EDY9 A0A1B6F437 E2BNY4 A0A0V0GB39 A0A1B6DB48 A0A1B6CGZ9 A0A0L7R9Y6 A0A1B6HYR9 A0A088ACZ8 A0A023EY28 A0A1S3H980 A0A293LUZ6 A0A1B6LCA4 A0A195CSU0 A0A151IYV7 E0VHV7 A0A151X778 A0A310SUW4 D6WZH5 A0A210QTT4 A0A158NWK4 F4WZ85 V4AS73 A0A195F7S5 A7S4R7 J9JVQ7 A0A177WCI4 F4PBJ7 A0A195AU82 A0A2H8TLU9 A0A3M7T215 A0A224Z6A9 E9I915 A0A1Y1MLM5 A0A026X1W0 A0A3L8DV12 L8HCQ5 A0A3S1A6Y9 A0A131YWV9 A0A1E1XC80 A0A1D2MPM0 A0A1Y1Y108 A0A075AUJ9 N6UPM9 A0A0A9W3L7 T1HJ73 A0A1W4XI64 A0A146LA62 A0A2S2QPV5 B7PS70 A0A3S3SC75 A0A196SFE6 A0A131XSM5 A0A2G8K9D4 A0A2B4RXG8 A0A1E1W4W7 A0A0B6ZFZ7 A0A1B6DFL6 A0A151M2B1 A0A1B6D868 A0A1U7RDZ6 A0A0L0HSE9 A0A1S8W8V7 U4L276 A0A026WYM1 A0A3L8D835 G3MFK5 A0A0Q3USM3 A0A023GNP6 I3M6D0 A0A0P5VX41 A0A3M6TI40 A0A0N7ZN89 A0A2A4JHI6
A0A212F817 A0A2H1WWY1 A0A0L7LAH1 A0A2J7R7Q0 A0A067RCE4 A0A2J7R7Q3 A0A2J7R7Q1 A0A2P8XTI4 A0A154PK67 A0A0J7L5J8 A0A3Q0J8T0 E2ANM1 A0A224XIL7 A0A0M9A428 A0A1Z5KY51 A0A2R5LL04 A0A2A3EDY9 A0A1B6F437 E2BNY4 A0A0V0GB39 A0A1B6DB48 A0A1B6CGZ9 A0A0L7R9Y6 A0A1B6HYR9 A0A088ACZ8 A0A023EY28 A0A1S3H980 A0A293LUZ6 A0A1B6LCA4 A0A195CSU0 A0A151IYV7 E0VHV7 A0A151X778 A0A310SUW4 D6WZH5 A0A210QTT4 A0A158NWK4 F4WZ85 V4AS73 A0A195F7S5 A7S4R7 J9JVQ7 A0A177WCI4 F4PBJ7 A0A195AU82 A0A2H8TLU9 A0A3M7T215 A0A224Z6A9 E9I915 A0A1Y1MLM5 A0A026X1W0 A0A3L8DV12 L8HCQ5 A0A3S1A6Y9 A0A131YWV9 A0A1E1XC80 A0A1D2MPM0 A0A1Y1Y108 A0A075AUJ9 N6UPM9 A0A0A9W3L7 T1HJ73 A0A1W4XI64 A0A146LA62 A0A2S2QPV5 B7PS70 A0A3S3SC75 A0A196SFE6 A0A131XSM5 A0A2G8K9D4 A0A2B4RXG8 A0A1E1W4W7 A0A0B6ZFZ7 A0A1B6DFL6 A0A151M2B1 A0A1B6D868 A0A1U7RDZ6 A0A0L0HSE9 A0A1S8W8V7 U4L276 A0A026WYM1 A0A3L8D835 G3MFK5 A0A0Q3USM3 A0A023GNP6 I3M6D0 A0A0P5VX41 A0A3M6TI40 A0A0N7ZN89 A0A2A4JHI6
EC Number
3.1.13.-
Pubmed
19121390
26919431
26354079
22118469
26227816
24845553
+ More
29403074 20798317 28528879 25474469 20566863 18362917 19820115 28812685 21347285 21719571 23254933 17615350 30375419 28797301 21282665 28004739 24508170 30249741 23375108 26830274 28503490 27289101 23932404 23537049 25401762 26823975 29023486 22293439 24068976 22216098 30382153
29403074 20798317 28528879 25474469 20566863 18362917 19820115 28812685 21347285 21719571 23254933 17615350 30375419 28797301 21282665 28004739 24508170 30249741 23375108 26830274 28503490 27289101 23932404 23537049 25401762 26823975 29023486 22293439 24068976 22216098 30382153
EMBL
BABH01008568
BABH01008569
RSAL01000081
RVE48606.1
NWSH01000001
PCG81122.1
+ More
KQ459606 KPI91649.1 KQ461198 KPJ06242.1 AGBW02009807 OWR49885.1 ODYU01011676 SOQ57573.1 JTDY01002063 KOB72201.1 NEVH01006727 PNF36862.1 KK852556 KDR21432.1 PNF36863.1 PNF36864.1 PYGN01001374 PSN35310.1 KQ434943 KZC12235.1 LBMM01000487 KMQ98242.1 GL441256 EFN64967.1 GFTR01004090 JAW12336.1 KQ435755 KOX76046.1 GFJQ02006924 JAW00046.1 GGLE01006067 MBY10193.1 KZ288285 PBC29502.1 GECZ01024832 JAS44937.1 GL449511 EFN82576.1 GECL01000966 JAP05158.1 GEDC01014385 JAS22913.1 GEDC01024522 JAS12776.1 KQ414620 KOC67680.1 GECU01027880 JAS79826.1 GBBI01004750 JAC13962.1 GFWV01007803 MAA32533.1 GEBQ01018658 JAT21319.1 KQ977305 KYN03705.1 KQ980735 KYN13751.1 DS235172 EEB12963.1 KQ982450 KYQ56227.1 KQ759903 OAD62005.1 KQ971372 EFA09703.1 NEDP02001925 OWF52139.1 ADTU01028122 GL888470 EGI60474.1 KB201304 ESO97720.1 KQ981756 KYN36114.1 DS469579 EDO41332.1 ABLF02022439 DS022301 OAJ37789.1 GL882892 EGF77459.1 KQ976741 KYM75585.1 GFXV01003332 MBW15137.1 REGN01000429 RNA41987.1 GFPF01010674 MAA21820.1 GL761667 EFZ22924.1 GEZM01027912 JAV86582.1 KK107024 EZA62295.1 QOIP01000004 RLU24257.1 KB007890 ELR22146.1 RQTK01000212 RUS84255.1 GEDV01005110 JAP83447.1 GFAC01002331 JAT96857.1 LJIJ01000726 ODM94983.1 MCFE01000310 ORX91701.1 KE561170 EPZ32187.1 APGK01022442 APGK01022443 APGK01022444 APGK01022445 KB740424 ENN80687.1 GBHO01044144 JAF99459.1 ACPB03017011 GDHC01013405 JAQ05224.1 GGMS01010357 MBY79560.1 ABJB010371410 DS777384 EEC09442.1 NCKU01001410 NCKU01001402 NCKU01001390 RWS12161.1 RWS12176.1 RWS12222.1 LXWW01000183 OAO15046.1 GEFM01006139 JAP69657.1 MRZV01000766 PIK44614.1 LSMT01000243 PFX22311.1 GDQN01009050 JAT82004.1 HACG01019916 CEK66781.1 GEDC01012886 JAS24412.1 AKHW03006780 KYO18664.1 GEDC01015405 JAS21893.1 KQ257451 KND03779.1 LYON01000053 OON10513.1 HF935286 CCX06373.1 KK107078 EZA60239.1 QOIP01000011 RLU16444.1 JO840656 AEO32273.1 LMAW01002292 KQK81594.1 GBBM01000843 JAC34575.1 AGTP01022054 GDIP01094940 JAM08775.1 RCHS01003537 RMX40991.1 GDIP01228825 JAI94576.1 NWSH01001365 PCG71535.1
KQ459606 KPI91649.1 KQ461198 KPJ06242.1 AGBW02009807 OWR49885.1 ODYU01011676 SOQ57573.1 JTDY01002063 KOB72201.1 NEVH01006727 PNF36862.1 KK852556 KDR21432.1 PNF36863.1 PNF36864.1 PYGN01001374 PSN35310.1 KQ434943 KZC12235.1 LBMM01000487 KMQ98242.1 GL441256 EFN64967.1 GFTR01004090 JAW12336.1 KQ435755 KOX76046.1 GFJQ02006924 JAW00046.1 GGLE01006067 MBY10193.1 KZ288285 PBC29502.1 GECZ01024832 JAS44937.1 GL449511 EFN82576.1 GECL01000966 JAP05158.1 GEDC01014385 JAS22913.1 GEDC01024522 JAS12776.1 KQ414620 KOC67680.1 GECU01027880 JAS79826.1 GBBI01004750 JAC13962.1 GFWV01007803 MAA32533.1 GEBQ01018658 JAT21319.1 KQ977305 KYN03705.1 KQ980735 KYN13751.1 DS235172 EEB12963.1 KQ982450 KYQ56227.1 KQ759903 OAD62005.1 KQ971372 EFA09703.1 NEDP02001925 OWF52139.1 ADTU01028122 GL888470 EGI60474.1 KB201304 ESO97720.1 KQ981756 KYN36114.1 DS469579 EDO41332.1 ABLF02022439 DS022301 OAJ37789.1 GL882892 EGF77459.1 KQ976741 KYM75585.1 GFXV01003332 MBW15137.1 REGN01000429 RNA41987.1 GFPF01010674 MAA21820.1 GL761667 EFZ22924.1 GEZM01027912 JAV86582.1 KK107024 EZA62295.1 QOIP01000004 RLU24257.1 KB007890 ELR22146.1 RQTK01000212 RUS84255.1 GEDV01005110 JAP83447.1 GFAC01002331 JAT96857.1 LJIJ01000726 ODM94983.1 MCFE01000310 ORX91701.1 KE561170 EPZ32187.1 APGK01022442 APGK01022443 APGK01022444 APGK01022445 KB740424 ENN80687.1 GBHO01044144 JAF99459.1 ACPB03017011 GDHC01013405 JAQ05224.1 GGMS01010357 MBY79560.1 ABJB010371410 DS777384 EEC09442.1 NCKU01001410 NCKU01001402 NCKU01001390 RWS12161.1 RWS12176.1 RWS12222.1 LXWW01000183 OAO15046.1 GEFM01006139 JAP69657.1 MRZV01000766 PIK44614.1 LSMT01000243 PFX22311.1 GDQN01009050 JAT82004.1 HACG01019916 CEK66781.1 GEDC01012886 JAS24412.1 AKHW03006780 KYO18664.1 GEDC01015405 JAS21893.1 KQ257451 KND03779.1 LYON01000053 OON10513.1 HF935286 CCX06373.1 KK107078 EZA60239.1 QOIP01000011 RLU16444.1 JO840656 AEO32273.1 LMAW01002292 KQK81594.1 GBBM01000843 JAC34575.1 AGTP01022054 GDIP01094940 JAM08775.1 RCHS01003537 RMX40991.1 GDIP01228825 JAI94576.1 NWSH01001365 PCG71535.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000235965 UP000027135 UP000245037 UP000076502 UP000036403 UP000079169 UP000000311 UP000053105 UP000242457 UP000008237 UP000053825 UP000005203 UP000085678 UP000078542 UP000078492 UP000009046 UP000075809 UP000007266 UP000242188 UP000005205 UP000007755 UP000030746 UP000078541 UP000001593 UP000007819 UP000077115 UP000007241 UP000078540 UP000276133 UP000053097 UP000279307 UP000011083 UP000271974 UP000094527 UP000193498 UP000030755 UP000019118 UP000015103 UP000192223 UP000001555 UP000285301 UP000078348 UP000230750 UP000225706 UP000050525 UP000189705 UP000053201 UP000189906 UP000018144 UP000051836 UP000005215 UP000275408
UP000037510 UP000235965 UP000027135 UP000245037 UP000076502 UP000036403 UP000079169 UP000000311 UP000053105 UP000242457 UP000008237 UP000053825 UP000005203 UP000085678 UP000078542 UP000078492 UP000009046 UP000075809 UP000007266 UP000242188 UP000005205 UP000007755 UP000030746 UP000078541 UP000001593 UP000007819 UP000077115 UP000007241 UP000078540 UP000276133 UP000053097 UP000279307 UP000011083 UP000271974 UP000094527 UP000193498 UP000030755 UP000019118 UP000015103 UP000192223 UP000001555 UP000285301 UP000078348 UP000230750 UP000225706 UP000050525 UP000189705 UP000053201 UP000189906 UP000018144 UP000051836 UP000005215 UP000275408
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A1U6USN9
H9JAQ7
A0A3S2NZN8
A0A2A4KBR8
A0A194PG21
A0A194QL30
+ More
A0A212F817 A0A2H1WWY1 A0A0L7LAH1 A0A2J7R7Q0 A0A067RCE4 A0A2J7R7Q3 A0A2J7R7Q1 A0A2P8XTI4 A0A154PK67 A0A0J7L5J8 A0A3Q0J8T0 E2ANM1 A0A224XIL7 A0A0M9A428 A0A1Z5KY51 A0A2R5LL04 A0A2A3EDY9 A0A1B6F437 E2BNY4 A0A0V0GB39 A0A1B6DB48 A0A1B6CGZ9 A0A0L7R9Y6 A0A1B6HYR9 A0A088ACZ8 A0A023EY28 A0A1S3H980 A0A293LUZ6 A0A1B6LCA4 A0A195CSU0 A0A151IYV7 E0VHV7 A0A151X778 A0A310SUW4 D6WZH5 A0A210QTT4 A0A158NWK4 F4WZ85 V4AS73 A0A195F7S5 A7S4R7 J9JVQ7 A0A177WCI4 F4PBJ7 A0A195AU82 A0A2H8TLU9 A0A3M7T215 A0A224Z6A9 E9I915 A0A1Y1MLM5 A0A026X1W0 A0A3L8DV12 L8HCQ5 A0A3S1A6Y9 A0A131YWV9 A0A1E1XC80 A0A1D2MPM0 A0A1Y1Y108 A0A075AUJ9 N6UPM9 A0A0A9W3L7 T1HJ73 A0A1W4XI64 A0A146LA62 A0A2S2QPV5 B7PS70 A0A3S3SC75 A0A196SFE6 A0A131XSM5 A0A2G8K9D4 A0A2B4RXG8 A0A1E1W4W7 A0A0B6ZFZ7 A0A1B6DFL6 A0A151M2B1 A0A1B6D868 A0A1U7RDZ6 A0A0L0HSE9 A0A1S8W8V7 U4L276 A0A026WYM1 A0A3L8D835 G3MFK5 A0A0Q3USM3 A0A023GNP6 I3M6D0 A0A0P5VX41 A0A3M6TI40 A0A0N7ZN89 A0A2A4JHI6
A0A212F817 A0A2H1WWY1 A0A0L7LAH1 A0A2J7R7Q0 A0A067RCE4 A0A2J7R7Q3 A0A2J7R7Q1 A0A2P8XTI4 A0A154PK67 A0A0J7L5J8 A0A3Q0J8T0 E2ANM1 A0A224XIL7 A0A0M9A428 A0A1Z5KY51 A0A2R5LL04 A0A2A3EDY9 A0A1B6F437 E2BNY4 A0A0V0GB39 A0A1B6DB48 A0A1B6CGZ9 A0A0L7R9Y6 A0A1B6HYR9 A0A088ACZ8 A0A023EY28 A0A1S3H980 A0A293LUZ6 A0A1B6LCA4 A0A195CSU0 A0A151IYV7 E0VHV7 A0A151X778 A0A310SUW4 D6WZH5 A0A210QTT4 A0A158NWK4 F4WZ85 V4AS73 A0A195F7S5 A7S4R7 J9JVQ7 A0A177WCI4 F4PBJ7 A0A195AU82 A0A2H8TLU9 A0A3M7T215 A0A224Z6A9 E9I915 A0A1Y1MLM5 A0A026X1W0 A0A3L8DV12 L8HCQ5 A0A3S1A6Y9 A0A131YWV9 A0A1E1XC80 A0A1D2MPM0 A0A1Y1Y108 A0A075AUJ9 N6UPM9 A0A0A9W3L7 T1HJ73 A0A1W4XI64 A0A146LA62 A0A2S2QPV5 B7PS70 A0A3S3SC75 A0A196SFE6 A0A131XSM5 A0A2G8K9D4 A0A2B4RXG8 A0A1E1W4W7 A0A0B6ZFZ7 A0A1B6DFL6 A0A151M2B1 A0A1B6D868 A0A1U7RDZ6 A0A0L0HSE9 A0A1S8W8V7 U4L276 A0A026WYM1 A0A3L8D835 G3MFK5 A0A0Q3USM3 A0A023GNP6 I3M6D0 A0A0P5VX41 A0A3M6TI40 A0A0N7ZN89 A0A2A4JHI6
PDB
3D45
E-value=6.41748e-18,
Score=219
Ontologies
GO
Topology
Subcellular location
Mitochondrion outer membrane
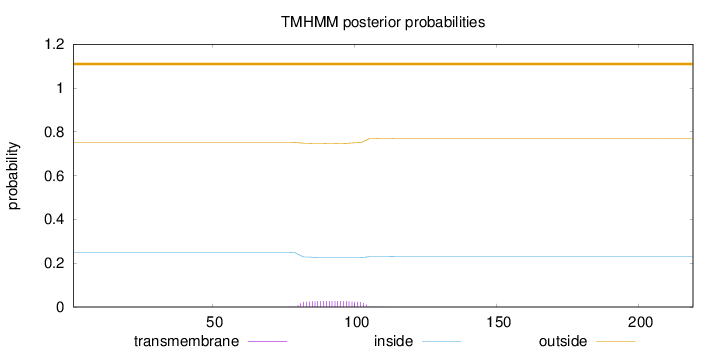
Length:
219
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.60494
Exp number, first 60 AAs:
0.00082
Total prob of N-in:
0.24901
outside
1 - 219
Population Genetic Test Statistics
Pi
270.47266
Theta
203.634444
Tajima's D
1.350013
CLR
0
CSRT
0.747612619369032
Interpretation
Uncertain
