Gene
KWMTBOMO05542
Pre Gene Modal
BGIBMGA006602
Annotation
PREDICTED:_poly(A)-specific_ribonuclease_PARN-like_domain-containing_protein_1_[Papilio_xuthus]
Full name
Pre-piRNA 3'-exonuclease trimmer
+ More
Poly(A)-specific ribonuclease PNLDC1
Poly(A)-specific ribonuclease PNLDC1
Alternative Name
PARN-like domain-containing protein 1 homolog
PARN-like domain-containing protein 1
Poly(A)-specific ribonuclease domain-containing protein 1
PARN-like domain-containing protein 1
Poly(A)-specific ribonuclease domain-containing protein 1
Location in the cell
PlasmaMembrane Reliability : 2.736
Sequence
CDS
ATGCCTGACCTGATTTTAACTTGCTCTTTCTATTTAAACATTCTCATATACAGAGACAAAAATGTTGAAGGAGCCAAAAATGCACCAATAGCAATACTGGAAGATAACTTAATAGCCTATTTACTGGGCTTCTTGCACGTAATAAAACTGCTAGAAACACACAAGAAGCCCATTGTAGGACACAACATGTTTTTGGATATGTTGTTCCTTCACAATCAATTCATAGGACCACTACCGGATAGTTATACAATGTTTAAAAAGAACATTAACAGTGTATTTCCAACTATATTTGATACTAAATATATATCGCATGCTATGAGTAAGAAACTCACTTTTTCTGAGTCTTGGAAATCAAATGCATTACAAGATTTGTATGAATTCTTCGCTGAAAGAAAATGTAAGAAACTTGAATATGGAATCAACATTGTACGACTGACCACACCATTTGATATTAAACAAAGCTATCATGAAGCCGGATGGGACGCTTACTGTTCAGGTTATTGCTTTATACGGCTCGGTCACTGGGCCGCCTGCGAGAACCGCGGCAAGTCTCACGTGATCGGACCCAGGGAGAAACTCGCAGCCCTAGCGCCTTACTGCAACAAGGTCAATGTTATAAGAGGTGGCGTTTCTTATATGAATTTCTCAGAAAACGATCCACCTCGGAACAGACCTGTCTTGTTGCACGTTAAATGTCTAAAAGACACCGTCATAGATGTTGAAAAGGTGGCGTCGCTACTGGGCAGCTTCGGTTCGATCGACATTAAGCCCTGTGGCAAGCGAGCCGCCTTGGTAGCGACCGGCGCACAGTTTATGGTAGACAAAATTCTGAAAACCTATGAAAACAACAGAGATTATAGAATTTCGAAATACAGTGTATACAAGCATTCAGTTGCAGGTCGCTTCGCTATCTGGAGCGGTTCGATTGTAACAGGCGGCCTGGCGTTATACTTAATTCATAAGAAATTTAAATCTATTTTGTTATAA
Protein
MPDLILTCSFYLNILIYRDKNVEGAKNAPIAILEDNLIAYLLGFLHVIKLLETHKKPIVGHNMFLDMLFLHNQFIGPLPDSYTMFKKNINSVFPTIFDTKYISHAMSKKLTFSESWKSNALQDLYEFFAERKCKKLEYGINIVRLTTPFDIKQSYHEAGWDAYCSGYCFIRLGHWAACENRGKSHVIGPREKLAALAPYCNKVNVIRGGVSYMNFSENDPPRNRPVLLHVKCLKDTVIDVEKVASLLGSFGSIDIKPCGKRAALVATGAQFMVDKILKTYENNRDYRISKYSVYKHSVAGRFAIWSGSIVTGGLALYLIHKKFKSILL
Summary
Description
3'-5' exonuclease that specifically cleaves precursor piRNAs (pre-piRNAs) at their 3' ends (PubMed:26919431). Trims pre-piRNAs to their mature size, a process required for piRNAs maturation and stabilization, and subsequent pre-piRNAs 2'-O-methylation (PubMed:26919431). The piRNA metabolic process mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and govern the methylation and subsequent repression of transposons (PubMed:26919431).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails (PubMed:27515512). Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development (PubMed:27515512). May act as a regulator of multipotency in embryonic stem cells (PubMed:27515512).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails (PubMed:27515512). Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development (PubMed:27515512). May act as a regulator of multipotency in embryonic stem cells (PubMed:27515512).
Catalytic Activity
Exonucleolytic cleavage of poly(A) to 5'-AMP.
Cofactor
Mg(2+)
Subunit
Interacts with Papi/Tdrkh; interaction takes place on the mitochondrial surface and recruits PNLDC1/trimmer to PIWI-bound pre-piRNAs.
Similarity
Belongs to the CAF1 family.
Keywords
Complete proteome
Exonuclease
Hydrolase
Magnesium
Membrane
Metal-binding
Mitochondrion
Mitochondrion outer membrane
Nuclease
Reference proteome
RNA-binding
RNA-mediated gene silencing
Transmembrane
Transmembrane helix
Endoplasmic reticulum
Nonsense-mediated mRNA decay
Feature
chain Pre-piRNA 3'-exonuclease trimmer
Uniprot
H9JAQ7
A0A1U6USN9
A0A2A4KBR8
A0A3S2NZN8
A0A194QL30
A0A2H1WWY1
+ More
A0A212F817 A0A194PG21 A0A1B6H5Y4 A0A1B6F437 A0A1B6LCA4 A0A2J7R7Q5 A0A1B6DB48 A0A1B6CGZ9 A0A1W4XI64 A0A2J7R7Q7 A0A2J7R7Q1 A0A2J7R7Q3 A0A067RCE4 A0A026X1W0 A0A088ACZ8 A0A2J7R7R9 A0A195CSU0 A0A151IYV7 A0A195F7S5 A0A151X778 A0A3L8DV12 A0A2P8XTI4 F4WZ85 A0A1S3H980 D6WZH5 E2ANM1 A0A1Y1MLM5 A0A2A3EDY9 A0A023EY28 A0A154PK67 A0A310SUW4 A0A0V0GB39 A0A146LA62 A0A0M9A428 A0A3S1A6Y9 A0A195AU82 A0A210QTT4 A0A2G8K9D4 V4AS73 A0A293LUZ6 A0A1Z5KY51 N6UPM9 R7T383 A0A094K7S6 A0A091S4J2 C3YTQ7 A0A2R5LL04 A0A091V3Z6 A0A093IWN0 E9I915 A0A091JSK6 A0A091KCV3 A0A452HZW9 A0A2I0MU41 A0A087QH11 A0A091WK52 A0A091UI22 A0A091PNH4 A0A091FXH2 A0A1V4JKZ1 A0A1V4JL15 T1KR32 A0A1E1XC80 A0A023GNP6 U3IHA7 A0A452HZS3 A0A452HZU7 A0A093H1K7 A0A093BYZ8 A0A091HQB3 A0A093FRT1 B2RXZ1 G3MFK5 A0A093PU64 A0A091RA35 A0A2K5QLT1 A0A087VF91 M3Y0M9 A0A091QHB6 R0K1Y6 H0ZME9 A0A2K6SBZ7 A0A091NAL1 A0A093R9F8 A0A3Q0DIW4 A0A131YWV9 A0A2R8ZR07 G2HGJ5 A0A2Y9KTU2
A0A212F817 A0A194PG21 A0A1B6H5Y4 A0A1B6F437 A0A1B6LCA4 A0A2J7R7Q5 A0A1B6DB48 A0A1B6CGZ9 A0A1W4XI64 A0A2J7R7Q7 A0A2J7R7Q1 A0A2J7R7Q3 A0A067RCE4 A0A026X1W0 A0A088ACZ8 A0A2J7R7R9 A0A195CSU0 A0A151IYV7 A0A195F7S5 A0A151X778 A0A3L8DV12 A0A2P8XTI4 F4WZ85 A0A1S3H980 D6WZH5 E2ANM1 A0A1Y1MLM5 A0A2A3EDY9 A0A023EY28 A0A154PK67 A0A310SUW4 A0A0V0GB39 A0A146LA62 A0A0M9A428 A0A3S1A6Y9 A0A195AU82 A0A210QTT4 A0A2G8K9D4 V4AS73 A0A293LUZ6 A0A1Z5KY51 N6UPM9 R7T383 A0A094K7S6 A0A091S4J2 C3YTQ7 A0A2R5LL04 A0A091V3Z6 A0A093IWN0 E9I915 A0A091JSK6 A0A091KCV3 A0A452HZW9 A0A2I0MU41 A0A087QH11 A0A091WK52 A0A091UI22 A0A091PNH4 A0A091FXH2 A0A1V4JKZ1 A0A1V4JL15 T1KR32 A0A1E1XC80 A0A023GNP6 U3IHA7 A0A452HZS3 A0A452HZU7 A0A093H1K7 A0A093BYZ8 A0A091HQB3 A0A093FRT1 B2RXZ1 G3MFK5 A0A093PU64 A0A091RA35 A0A2K5QLT1 A0A087VF91 M3Y0M9 A0A091QHB6 R0K1Y6 H0ZME9 A0A2K6SBZ7 A0A091NAL1 A0A093R9F8 A0A3Q0DIW4 A0A131YWV9 A0A2R8ZR07 G2HGJ5 A0A2Y9KTU2
EC Number
3.1.13.-
3.1.13.4
3.1.13.4
Pubmed
19121390
26919431
26354079
22118469
24845553
24508170
+ More
30249741 29403074 21719571 18362917 19820115 20798317 28004739 25474469 26823975 28812685 29023486 23254933 28528879 23537049 18563158 21282665 28562605 23371554 28503490 23749191 19468303 15489334 27515512 22216098 20360741 26830274 22722832 21484476
30249741 29403074 21719571 18362917 19820115 20798317 28004739 25474469 26823975 28812685 29023486 23254933 28528879 23537049 18563158 21282665 28562605 23371554 28503490 23749191 19468303 15489334 27515512 22216098 20360741 26830274 22722832 21484476
EMBL
BABH01008568
BABH01008569
NWSH01000001
PCG81122.1
RSAL01000081
RVE48606.1
+ More
KQ461198 KPJ06242.1 ODYU01011676 SOQ57573.1 AGBW02009807 OWR49885.1 KQ459606 KPI91649.1 GECU01037591 JAS70115.1 GECZ01024832 JAS44937.1 GEBQ01018658 JAT21319.1 NEVH01006727 PNF36867.1 GEDC01014385 JAS22913.1 GEDC01024522 JAS12776.1 PNF36865.1 PNF36864.1 PNF36863.1 KK852556 KDR21432.1 KK107024 EZA62295.1 PNF36866.1 KQ977305 KYN03705.1 KQ980735 KYN13751.1 KQ981756 KYN36114.1 KQ982450 KYQ56227.1 QOIP01000004 RLU24257.1 PYGN01001374 PSN35310.1 GL888470 EGI60474.1 KQ971372 EFA09703.1 GL441256 EFN64967.1 GEZM01027912 JAV86582.1 KZ288285 PBC29502.1 GBBI01004750 JAC13962.1 KQ434943 KZC12235.1 KQ759903 OAD62005.1 GECL01000966 JAP05158.1 GDHC01013405 JAQ05224.1 KQ435755 KOX76046.1 RQTK01000212 RUS84255.1 KQ976741 KYM75585.1 NEDP02001925 OWF52139.1 MRZV01000766 PIK44614.1 KB201304 ESO97720.1 GFWV01007803 MAA32533.1 GFJQ02006924 JAW00046.1 APGK01022442 APGK01022443 APGK01022444 APGK01022445 KB740424 ENN80687.1 AMQN01003698 KB312525 ELT87068.1 KL341697 KFZ53302.1 KK941262 KFQ50871.1 GG666552 EEN56184.1 GGLE01006067 MBY10193.1 KL410630 KFQ98036.1 KK600886 KFW06043.1 GL761667 EFZ22924.1 KK501404 KFP14611.1 KK739865 KFP38394.1 AKCR02000002 PKK33198.1 KL225581 KFM00515.1 KK735662 KFR15283.1 KK452852 KFQ74597.1 KK662111 KFQ08896.1 KL447472 KFO73801.1 LSYS01006902 OPJ72849.1 OPJ72850.1 CAEY01000383 GFAC01002331 JAT96857.1 GBBM01000843 JAC34575.1 ADON01047579 KL217258 KFV75551.1 KL230267 KFV05207.1 KL217729 KFO98101.1 KK395402 KFV57036.1 AL589878 BC158033 JO840656 AEO32273.1 KL670888 KFW80343.1 KK811878 KFQ36117.1 KL491093 KFO11283.1 AEYP01076172 KK675958 KFQ08909.1 KB742815 EOB03981.1 ABQF01000072 ABQF01000073 KK844906 KFP86718.1 KL444849 KFW95165.1 GEDV01005110 JAP83447.1 AJFE02021429 AK305859 BAK62853.1
KQ461198 KPJ06242.1 ODYU01011676 SOQ57573.1 AGBW02009807 OWR49885.1 KQ459606 KPI91649.1 GECU01037591 JAS70115.1 GECZ01024832 JAS44937.1 GEBQ01018658 JAT21319.1 NEVH01006727 PNF36867.1 GEDC01014385 JAS22913.1 GEDC01024522 JAS12776.1 PNF36865.1 PNF36864.1 PNF36863.1 KK852556 KDR21432.1 KK107024 EZA62295.1 PNF36866.1 KQ977305 KYN03705.1 KQ980735 KYN13751.1 KQ981756 KYN36114.1 KQ982450 KYQ56227.1 QOIP01000004 RLU24257.1 PYGN01001374 PSN35310.1 GL888470 EGI60474.1 KQ971372 EFA09703.1 GL441256 EFN64967.1 GEZM01027912 JAV86582.1 KZ288285 PBC29502.1 GBBI01004750 JAC13962.1 KQ434943 KZC12235.1 KQ759903 OAD62005.1 GECL01000966 JAP05158.1 GDHC01013405 JAQ05224.1 KQ435755 KOX76046.1 RQTK01000212 RUS84255.1 KQ976741 KYM75585.1 NEDP02001925 OWF52139.1 MRZV01000766 PIK44614.1 KB201304 ESO97720.1 GFWV01007803 MAA32533.1 GFJQ02006924 JAW00046.1 APGK01022442 APGK01022443 APGK01022444 APGK01022445 KB740424 ENN80687.1 AMQN01003698 KB312525 ELT87068.1 KL341697 KFZ53302.1 KK941262 KFQ50871.1 GG666552 EEN56184.1 GGLE01006067 MBY10193.1 KL410630 KFQ98036.1 KK600886 KFW06043.1 GL761667 EFZ22924.1 KK501404 KFP14611.1 KK739865 KFP38394.1 AKCR02000002 PKK33198.1 KL225581 KFM00515.1 KK735662 KFR15283.1 KK452852 KFQ74597.1 KK662111 KFQ08896.1 KL447472 KFO73801.1 LSYS01006902 OPJ72849.1 OPJ72850.1 CAEY01000383 GFAC01002331 JAT96857.1 GBBM01000843 JAC34575.1 ADON01047579 KL217258 KFV75551.1 KL230267 KFV05207.1 KL217729 KFO98101.1 KK395402 KFV57036.1 AL589878 BC158033 JO840656 AEO32273.1 KL670888 KFW80343.1 KK811878 KFQ36117.1 KL491093 KFO11283.1 AEYP01076172 KK675958 KFQ08909.1 KB742815 EOB03981.1 ABQF01000072 ABQF01000073 KK844906 KFP86718.1 KL444849 KFW95165.1 GEDV01005110 JAP83447.1 AJFE02021429 AK305859 BAK62853.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000235965 UP000192223 UP000027135 UP000053097 UP000005203 UP000078542 UP000078492 UP000078541 UP000075809 UP000279307 UP000245037 UP000007755 UP000085678 UP000007266 UP000000311 UP000242457 UP000076502 UP000053105 UP000271974 UP000078540 UP000242188 UP000230750 UP000030746 UP000019118 UP000014760 UP000001554 UP000053283 UP000053119 UP000291020 UP000053872 UP000053286 UP000053605 UP000053760 UP000190648 UP000015104 UP000016666 UP000053875 UP000054308 UP000000589 UP000053258 UP000233040 UP000000715 UP000007754 UP000233220 UP000189704 UP000240080 UP000248482
UP000235965 UP000192223 UP000027135 UP000053097 UP000005203 UP000078542 UP000078492 UP000078541 UP000075809 UP000279307 UP000245037 UP000007755 UP000085678 UP000007266 UP000000311 UP000242457 UP000076502 UP000053105 UP000271974 UP000078540 UP000242188 UP000230750 UP000030746 UP000019118 UP000014760 UP000001554 UP000053283 UP000053119 UP000291020 UP000053872 UP000053286 UP000053605 UP000053760 UP000190648 UP000015104 UP000016666 UP000053875 UP000054308 UP000000589 UP000053258 UP000233040 UP000000715 UP000007754 UP000233220 UP000189704 UP000240080 UP000248482
Interpro
Gene 3D
ProteinModelPortal
H9JAQ7
A0A1U6USN9
A0A2A4KBR8
A0A3S2NZN8
A0A194QL30
A0A2H1WWY1
+ More
A0A212F817 A0A194PG21 A0A1B6H5Y4 A0A1B6F437 A0A1B6LCA4 A0A2J7R7Q5 A0A1B6DB48 A0A1B6CGZ9 A0A1W4XI64 A0A2J7R7Q7 A0A2J7R7Q1 A0A2J7R7Q3 A0A067RCE4 A0A026X1W0 A0A088ACZ8 A0A2J7R7R9 A0A195CSU0 A0A151IYV7 A0A195F7S5 A0A151X778 A0A3L8DV12 A0A2P8XTI4 F4WZ85 A0A1S3H980 D6WZH5 E2ANM1 A0A1Y1MLM5 A0A2A3EDY9 A0A023EY28 A0A154PK67 A0A310SUW4 A0A0V0GB39 A0A146LA62 A0A0M9A428 A0A3S1A6Y9 A0A195AU82 A0A210QTT4 A0A2G8K9D4 V4AS73 A0A293LUZ6 A0A1Z5KY51 N6UPM9 R7T383 A0A094K7S6 A0A091S4J2 C3YTQ7 A0A2R5LL04 A0A091V3Z6 A0A093IWN0 E9I915 A0A091JSK6 A0A091KCV3 A0A452HZW9 A0A2I0MU41 A0A087QH11 A0A091WK52 A0A091UI22 A0A091PNH4 A0A091FXH2 A0A1V4JKZ1 A0A1V4JL15 T1KR32 A0A1E1XC80 A0A023GNP6 U3IHA7 A0A452HZS3 A0A452HZU7 A0A093H1K7 A0A093BYZ8 A0A091HQB3 A0A093FRT1 B2RXZ1 G3MFK5 A0A093PU64 A0A091RA35 A0A2K5QLT1 A0A087VF91 M3Y0M9 A0A091QHB6 R0K1Y6 H0ZME9 A0A2K6SBZ7 A0A091NAL1 A0A093R9F8 A0A3Q0DIW4 A0A131YWV9 A0A2R8ZR07 G2HGJ5 A0A2Y9KTU2
A0A212F817 A0A194PG21 A0A1B6H5Y4 A0A1B6F437 A0A1B6LCA4 A0A2J7R7Q5 A0A1B6DB48 A0A1B6CGZ9 A0A1W4XI64 A0A2J7R7Q7 A0A2J7R7Q1 A0A2J7R7Q3 A0A067RCE4 A0A026X1W0 A0A088ACZ8 A0A2J7R7R9 A0A195CSU0 A0A151IYV7 A0A195F7S5 A0A151X778 A0A3L8DV12 A0A2P8XTI4 F4WZ85 A0A1S3H980 D6WZH5 E2ANM1 A0A1Y1MLM5 A0A2A3EDY9 A0A023EY28 A0A154PK67 A0A310SUW4 A0A0V0GB39 A0A146LA62 A0A0M9A428 A0A3S1A6Y9 A0A195AU82 A0A210QTT4 A0A2G8K9D4 V4AS73 A0A293LUZ6 A0A1Z5KY51 N6UPM9 R7T383 A0A094K7S6 A0A091S4J2 C3YTQ7 A0A2R5LL04 A0A091V3Z6 A0A093IWN0 E9I915 A0A091JSK6 A0A091KCV3 A0A452HZW9 A0A2I0MU41 A0A087QH11 A0A091WK52 A0A091UI22 A0A091PNH4 A0A091FXH2 A0A1V4JKZ1 A0A1V4JL15 T1KR32 A0A1E1XC80 A0A023GNP6 U3IHA7 A0A452HZS3 A0A452HZU7 A0A093H1K7 A0A093BYZ8 A0A091HQB3 A0A093FRT1 B2RXZ1 G3MFK5 A0A093PU64 A0A091RA35 A0A2K5QLT1 A0A087VF91 M3Y0M9 A0A091QHB6 R0K1Y6 H0ZME9 A0A2K6SBZ7 A0A091NAL1 A0A093R9F8 A0A3Q0DIW4 A0A131YWV9 A0A2R8ZR07 G2HGJ5 A0A2Y9KTU2
PDB
3D45
E-value=8.88738e-19,
Score=229
Ontologies
GO
Topology
Subcellular location
Mitochondrion outer membrane
Endoplasmic reticulum membrane Localizes mainly in the endoplasmic reticulum. With evidence from 5 publications.
Endoplasmic reticulum membrane Localizes mainly in the endoplasmic reticulum. With evidence from 5 publications.
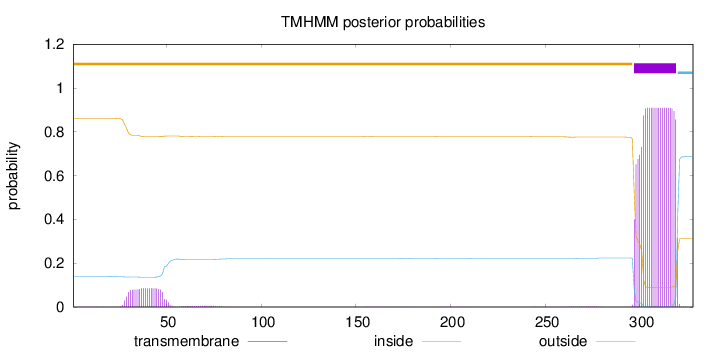
Length:
328
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.67408
Exp number, first 60 AAs:
1.75779
Total prob of N-in:
0.13807
outside
1 - 296
TMhelix
297 - 319
inside
320 - 328
Population Genetic Test Statistics
Pi
287.11442
Theta
197.855583
Tajima's D
0.665436
CLR
0.197957
CSRT
0.558522073896305
Interpretation
Uncertain
