Gene
KWMTBOMO05541 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006603
Annotation
sarco/endoplasmic_reticulum_calcium_ATPase_[Bombyx_mori]
Full name
Calcium-transporting ATPase
+ More
Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type
Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type
Alternative Name
Calcium pump
Calcium ATPase at 60A
Calcium ATPase at 60A
Location in the cell
PlasmaMembrane Reliability : 2.976
Sequence
CDS
ATGGAGGACGCTCACACGAAATCCGTGGAAGAAGTCTTAAAATATTTTGGCACAGACCCAGACAAAGGCCTTAGTCCAGACCAAATAAAAAGGAACCAAGAAAAATATGGACCCAATGAACTGCCTACGGAGGAAGGCAAAAGTATATGGCAGTTAGTCTTGGAACAATTCGACGACCTCTTAGTCAAGATTTTGCTGTTAGCTGCTATTATTTCATTCGTATTAGCTTTATTTGAAGAGCACGAAGATGCATTCTCAGCCTTCGTTGAACCTTTCGTTATTTTACTAATTCTTATTGCTAACGCTGTAGTAGGAGTATGGCAGGAAAGAAACGCCGAATCTGCCATCGAAGCTTTAAAAGAATACGAACCTGAAATGGGTAAAGTCATAAGAGGAGACAAATCTGGAGTACAAAAAATCCGTGCCAAGGAAATTGTTCCCGGGGACGTTGTTGAAGTGTCCGTTGGTGACAAGATCCCTGCTGACATTCGCCTTATCAAAATCTACTCCACCACAATCCGTATCGATCAGTCTATCCTGACTGGTGAATCTGTCTCGGTCATCAAACACACCGACCCCATTCCTGACCCCCGCGCCGTGAACCAAGACAAGAAGAACATTCTCTTCTCTGGTACTAATGTCGCAGCCGGTAAAGCTCGCGGTATCGTCATCGGTACTGGACTCAACACTGCCATCGGTAAGATCCGTACTGAAATGTCAGAAACCGAAGAAATCAAAACTCCTCTCCAACAGAAACTTGATGAGTTTGGTGAGCAGCTCTCTAAAGTAATCTCAGTTATCTGTGTCGCTGTGTGGGCTATTAACATCGGACACTTTAACGACCCCGCTCACGGTGGAAGTTGGATCAAGGGCGCCGTCTACTACTTCAAAATTGCCGTCGCTTTAGCCGTCGCTGCCATCCCTGAAGGTCTTCCGGCCGTCATCACCACTTGTCTCGCTTTGGGTACCAGAAGAATGGCTAAGAAAAACGCTATCGTAAGGTCTCTACCGTCAGTCGAAACCCTTGGCTGCACATCCGTCATCTGCTCCGACAAGACCGGCACACTGACCACTAACCAAATGTCCGTATCGCGCATGTTCATCTTTGAAAAGATCGAAGGTGGCGACAGCAGCTTCCTCGAGTTTGAAATCACTGGCTCAACTTATGAACCAATCGGCGACGTTTATCTGAAAGGACAGAAAGTTAAGGCTGCCGAATTTGATGCTCTTCACGAGATCGGTACCATCTGCGTAATGTGCAATGACTCCGCTATTGATTTCAACGAATTCAAACAGGCGTTCGAAAAGGTCGGCGAAGCTACCGAGACTGCTCTGATTGTTCTTGCTGAAAAAATGAATCCCTTCAATGTTCCTAAGACTGGCTTAGATCGCCGATCCTCTGCTATTGTTGTGCGTCAAGAAATTGAGACCAAGTGGAAGAAAGAATTCACTCTCGAGTTCTCTCGTGACAGGAAATCTATGTCGACATACTGCACACCCCTCAAACCCTCGCGCCTCGGCAACGGACCCAAACTATTTGTCAAGGGAGCACCCGAAGGTGTACTCGAACGTTGCACACACGCTCGCGTCGGCACCAGCAAGGTTCCTCTGACCACGACCCTGAAGAACCGCATCTTGGACCTGACCCGCCAGTACGGTACTGGTCGCGATACGCTGCGTTGCTTGGCCCTGGCCACGGCCGATAACCCCCTCAAACCCGACGAAATGGATCTTGGTGACTCCACCAAATTCTACACTTACGAAGTCAACCTTACGTTCGTCGGAGTCGTCGGCATGTTGGACCCCCCTCGTAAAGAAGTTTTCGACTCGATCGTTCGTTGTCGCGCTGCCGGTATTCGCGTAATCGTCATCACCGGTGACAACAAGTCCACTGCCGAAGCCATCTGCAGGCGTATCGGTGTATTCGGAGAGGACGAAGATACCACTGGCAAATCTTTCTCCGGTCGCGAGTTCGATGACCTGCCCATCGCCGAGCAGCGCAGCGCCTGCGCCAAGGCTCGCCTGTTCTCGCGCGTGGAGCCCGCGCACAAGTCTAAGATCGTAGAGTACCTGCAGAGCATGAACGAGATCTCCGCCATGACTGGTGACGGTGTGAATGACGCACCGGCTCTGAAGAAGGCCGAGATTGGTATTGCCATGGGATCCGGCACGGCCGTCGCGAAGTCCGCCGCCGAGATGGTGTTGGCCGACGACAACTTCTCCTCCATCGTCGCTGCTGTTGAGGAAGGTCGCGCCATTTACAACAACATGAAGCAGTTCATCCGCTATCTGATCTCATCCAACATCGGTGAGGTGGTCTCGATCTTCTTGACTGCCGCTCTGGGTCTGCCCGAAGCTCTGATCCCCGTCCAACTGCTGTGGGTGAACCTGGTCACTGACGGTCTGCCCGCTACCGCCCTCGGATTCAATCCTCCCGATCTCGACATCATGGACAAGCCCCCTCGCAAGGCTGACGAAGGTCTCATCTCCGGATGGCTGTTCTTCAGATACATGGCTATCGGTGGCTACGTAGGTGCGGCCACAGTCGGCGCTGCCTCCTGGTGGTTCATGTACTCCCCGTACGGACCTCAAATGACATACTGGCAACTCACTCATCACTTACAATGTATCAGCGGTGGTGATGAATTTAAGGGTGTTGACTGCAAAGTGTTCACTGACCCTCACCCGATGACGATGGCTCTCTCAGTGCTGGTCACAATCGAGATGTTAAACGCCATGAACAGCTTGTCTGAGAACCAGTCTCTGGTCACCATGCCGCCCTGGTCAAACTTGTGGCTCGTCGGCTCCATGGCGCTCTCCTTCACTCTTCATTTCGTCATCCTCTACGTCGAGGTTCTTTCGGCCGTGTTCCAAGTGACGCCGCTGTCGCTCGACGAGTGGGTAACCGTAATGAAGTTCTCAGTACCCGTGGTACTGCTGGATGAAGTGCTCAAATTCGTCGCGCGCAAGATCTCCGATGCGAACGATGTGGTCGTTGACAAGTGGTAA
Protein
MEDAHTKSVEEVLKYFGTDPDKGLSPDQIKRNQEKYGPNELPTEEGKSIWQLVLEQFDDLLVKILLLAAIISFVLALFEEHEDAFSAFVEPFVILLILIANAVVGVWQERNAESAIEALKEYEPEMGKVIRGDKSGVQKIRAKEIVPGDVVEVSVGDKIPADIRLIKIYSTTIRIDQSILTGESVSVIKHTDPIPDPRAVNQDKKNILFSGTNVAAGKARGIVIGTGLNTAIGKIRTEMSETEEIKTPLQQKLDEFGEQLSKVISVICVAVWAINIGHFNDPAHGGSWIKGAVYYFKIAVALAVAAIPEGLPAVITTCLALGTRRMAKKNAIVRSLPSVETLGCTSVICSDKTGTLTTNQMSVSRMFIFEKIEGGDSSFLEFEITGSTYEPIGDVYLKGQKVKAAEFDALHEIGTICVMCNDSAIDFNEFKQAFEKVGEATETALIVLAEKMNPFNVPKTGLDRRSSAIVVRQEIETKWKKEFTLEFSRDRKSMSTYCTPLKPSRLGNGPKLFVKGAPEGVLERCTHARVGTSKVPLTTTLKNRILDLTRQYGTGRDTLRCLALATADNPLKPDEMDLGDSTKFYTYEVNLTFVGVVGMLDPPRKEVFDSIVRCRAAGIRVIVITGDNKSTAEAICRRIGVFGEDEDTTGKSFSGREFDDLPIAEQRSACAKARLFSRVEPAHKSKIVEYLQSMNEISAMTGDGVNDAPALKKAEIGIAMGSGTAVAKSAAEMVLADDNFSSIVAAVEEGRAIYNNMKQFIRYLISSNIGEVVSIFLTAALGLPEALIPVQLLWVNLVTDGLPATALGFNPPDLDIMDKPPRKADEGLISGWLFFRYMAIGGYVGAATVGAASWWFMYSPYGPQMTYWQLTHHLQCISGGDEFKGVDCKVFTDPHPMTMALSVLVTIEMLNAMNSLSENQSLVTMPPWSNLWLVGSMALSFTLHFVILYVEVLSAVFQVTPLSLDEWVTVMKFSVPVVLLDEVLKFVARKISDANDVVVDKW
Summary
Description
This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium.
Catalytic Activity
ATP + Ca(2+)(in) + H2O = ADP + Ca(2+)(out) + H(+) + phosphate
Subunit
Interacts with SclA and SclB.
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Keywords
Alternative splicing
ATP-binding
Calcium
Calcium transport
Complete proteome
Endoplasmic reticulum
Ion transport
Magnesium
Membrane
Metal-binding
Nucleotide-binding
Phosphoprotein
Reference proteome
Sarcoplasmic reticulum
Translocase
Transmembrane
Transmembrane helix
Transport
Feature
chain Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type
splice variant In isoform A.
splice variant In isoform A.
Uniprot
C7AQP4
A0A2A4KA69
A0A3S2NIS6
A0A194QLJ6
O96696
A0A2A4KB22
+ More
A0A2H1W1G5 A0A1L8DF78 H9JAQ8 A0A212EY66 A0A0C9QYZ2 A0A0C9R823 A0A1L8DF68 A0A1B6JB05 A0A1B6K2B5 A0A385MDB2 A0A1B6E9S2 A0A224X6B8 D6WZH8 K7IPT6 A0A1B6CRW6 A0A154PK50 A0A1B6EA81 A0A069DXH6 A0A1B0G7F1 A0A2J7R7V8 A0A151IYX4 A0A023F5T4 A0A195F7F3 A0A195ATM9 A0A158P1I0 A0A195CU94 A0A0P4VR76 A0A1S4FE53 Q175R5 Q175R4 A0A0L0CN78 A0A067REG0 A0A151X776 A0A1Y1N0V8 A0A1W4X7R1 A0A2J7R7U2 A0A0K8S6S5 A0A088ACZ9 A0A0A1X082 T1DHY9 Q175R3 A0A1A9XZK2 A0A1B0BYV5 A0A1A9W106 T1PE27 A0A2J7R7U7 A0A1A9ZUZ6 A0A026X1Y8 A0A0V0G6F0 A0A1W4XH71 A0A0L7RAA5 A0A0K8UMR7 A0A034VML4 A0A2J7R7U1 V9IA48 A0A1Y1N483 V9I895 Q7PPA5-3 A0A1A9VNL8 A0A1Y1N586 A0A1W4X7X2 Q7PPA5-2 A0A0A1XTC4 B4LNZ0 U5EVU1 A0A0A1X2W8 A0A336KD28 W8AZ56 A0A084WL52 A0A1I8QEW5 B3MCM7 E0W0H6 Q7PPA5 A0A1I8QEQ1 B4MRN4 A0A2M3YXR9 A0A2M4CSZ5 A0A2S2QCR1 A0A034VKP1 A0A0Q9W4D5 B4PAA1 A0A2M3ZZM7 W8BAL8 B4KMB9 P22700-2 W8AV60 B3NQ25 A0A2M3ZZP9 A0A0Q9W432 A0A0J9RK49 B4I8W7 A0A182YKV8 A0A1W4V0L4 A0A1Q3FL77
A0A2H1W1G5 A0A1L8DF78 H9JAQ8 A0A212EY66 A0A0C9QYZ2 A0A0C9R823 A0A1L8DF68 A0A1B6JB05 A0A1B6K2B5 A0A385MDB2 A0A1B6E9S2 A0A224X6B8 D6WZH8 K7IPT6 A0A1B6CRW6 A0A154PK50 A0A1B6EA81 A0A069DXH6 A0A1B0G7F1 A0A2J7R7V8 A0A151IYX4 A0A023F5T4 A0A195F7F3 A0A195ATM9 A0A158P1I0 A0A195CU94 A0A0P4VR76 A0A1S4FE53 Q175R5 Q175R4 A0A0L0CN78 A0A067REG0 A0A151X776 A0A1Y1N0V8 A0A1W4X7R1 A0A2J7R7U2 A0A0K8S6S5 A0A088ACZ9 A0A0A1X082 T1DHY9 Q175R3 A0A1A9XZK2 A0A1B0BYV5 A0A1A9W106 T1PE27 A0A2J7R7U7 A0A1A9ZUZ6 A0A026X1Y8 A0A0V0G6F0 A0A1W4XH71 A0A0L7RAA5 A0A0K8UMR7 A0A034VML4 A0A2J7R7U1 V9IA48 A0A1Y1N483 V9I895 Q7PPA5-3 A0A1A9VNL8 A0A1Y1N586 A0A1W4X7X2 Q7PPA5-2 A0A0A1XTC4 B4LNZ0 U5EVU1 A0A0A1X2W8 A0A336KD28 W8AZ56 A0A084WL52 A0A1I8QEW5 B3MCM7 E0W0H6 Q7PPA5 A0A1I8QEQ1 B4MRN4 A0A2M3YXR9 A0A2M4CSZ5 A0A2S2QCR1 A0A034VKP1 A0A0Q9W4D5 B4PAA1 A0A2M3ZZM7 W8BAL8 B4KMB9 P22700-2 W8AV60 B3NQ25 A0A2M3ZZP9 A0A0Q9W432 A0A0J9RK49 B4I8W7 A0A182YKV8 A0A1W4V0L4 A0A1Q3FL77
EC Number
7.2.2.10
Pubmed
26354079
9528669
19121390
22118469
18362917
19820115
+ More
20075255 26334808 25474469 21347285 27129103 17510324 26108605 24845553 28004739 26823975 25830018 24330624 24508170 30249741 25348373 12364791 17994087 24495485 24438588 20566863 17550304 2148477 10731132 12537572 12537569 2557235 18327897 23970561 18057021 22936249 25244985
20075255 26334808 25474469 21347285 27129103 17510324 26108605 24845553 28004739 26823975 25830018 24330624 24508170 30249741 25348373 12364791 17994087 24495485 24438588 20566863 17550304 2148477 10731132 12537572 12537569 2557235 18327897 23970561 18057021 22936249 25244985
EMBL
FJ586237
ACU25861.1
NWSH01000001
PCG81121.1
RSAL01000081
RVE48605.1
+ More
KQ461198 KPJ06244.1 AF115572 AAD09820.1 PCG81115.1 PCG81114.1 ODYU01005735 SOQ46877.1 GFDF01009090 JAV04994.1 BABH01008562 AGBW02011611 OWR46425.1 GBYB01007896 GBYB01009024 JAG77663.1 JAG78791.1 GBYB01009022 JAG78789.1 GFDF01009084 JAV05000.1 GECU01011379 GECU01000717 JAS96327.1 JAT06990.1 GECU01002138 GECU01001496 JAT05569.1 JAT06211.1 MG271936 AYA58203.1 GEDC01002653 JAS34645.1 GFTR01008430 JAW07996.1 KQ971372 EFA10430.1 GEDC01021233 JAS16065.1 KQ434943 KZC12236.1 GEDC01026479 GEDC01022485 GEDC01002459 JAS10819.1 JAS14813.1 JAS34839.1 GBGD01000289 JAC88600.1 CCAG010021567 NEVH01006727 PNF36906.1 KQ980735 KYN13754.1 GBBI01002172 JAC16540.1 KQ981756 KYN36112.1 KQ976741 KYM75583.1 ADTU01006654 ADTU01006655 ADTU01006656 ADTU01006657 KQ977305 KYN03709.1 GDKW01001061 JAI55534.1 CH477396 EAT41828.1 EAT41827.1 JRES01000153 KNC33775.1 KK852556 KDR21433.1 KQ982450 KYQ56223.1 GEZM01015647 GEZM01015646 JAV91494.1 PNF36909.1 GBRD01016953 GDHC01003390 JAG48874.1 JAQ15239.1 GBXI01012040 GBXI01009573 JAD02252.1 JAD04719.1 GALA01001251 JAA93601.1 EAT41829.1 JXJN01022846 KA647017 AFP61646.1 PNF36908.1 KK107024 QOIP01000004 EZA62297.1 RLU24058.1 GECL01002565 JAP03559.1 KQ414620 KOC67681.1 GDHF01024541 JAI27773.1 GAKP01015902 JAC43050.1 PNF36907.1 JR036722 JR036724 AEY57349.1 AEY57351.1 GEZM01015645 JAV91495.1 JR036721 JR036723 AEY57348.1 AEY57350.1 AAAB01008960 GEZM01015644 JAV91496.1 GBXI01000062 JAD14230.1 CH940648 EDW61159.2 GANO01000863 JAB59008.1 GBXI01009274 GBXI01005620 JAD05018.1 JAD08672.1 UFQS01000224 UFQT01000224 SSX01628.1 SSX22008.1 GAMC01016417 JAB90138.1 ATLV01024183 KE525350 KFB50946.1 CH902619 EDV36261.1 AAZO01006743 DS235860 EEB19132.1 CH963850 EDW74773.1 GGFM01000321 MBW21072.1 GGFL01004262 MBW68440.1 GGMS01006323 MBY75526.1 GAKP01015903 JAC43049.1 KRF79821.1 CM000158 EDW92427.1 GGFK01000628 MBW33949.1 GAMC01016419 JAB90136.1 CH933808 EDW09807.1 M62892 AE013599 AY058465 X17472 GAMC01016418 JAB90137.1 CH954179 EDV56898.1 GGFK01000631 MBW33952.1 KRF79820.1 CM002911 KMY96172.1 CH480824 EDW57042.1 GFDL01006802 JAV28243.1
KQ461198 KPJ06244.1 AF115572 AAD09820.1 PCG81115.1 PCG81114.1 ODYU01005735 SOQ46877.1 GFDF01009090 JAV04994.1 BABH01008562 AGBW02011611 OWR46425.1 GBYB01007896 GBYB01009024 JAG77663.1 JAG78791.1 GBYB01009022 JAG78789.1 GFDF01009084 JAV05000.1 GECU01011379 GECU01000717 JAS96327.1 JAT06990.1 GECU01002138 GECU01001496 JAT05569.1 JAT06211.1 MG271936 AYA58203.1 GEDC01002653 JAS34645.1 GFTR01008430 JAW07996.1 KQ971372 EFA10430.1 GEDC01021233 JAS16065.1 KQ434943 KZC12236.1 GEDC01026479 GEDC01022485 GEDC01002459 JAS10819.1 JAS14813.1 JAS34839.1 GBGD01000289 JAC88600.1 CCAG010021567 NEVH01006727 PNF36906.1 KQ980735 KYN13754.1 GBBI01002172 JAC16540.1 KQ981756 KYN36112.1 KQ976741 KYM75583.1 ADTU01006654 ADTU01006655 ADTU01006656 ADTU01006657 KQ977305 KYN03709.1 GDKW01001061 JAI55534.1 CH477396 EAT41828.1 EAT41827.1 JRES01000153 KNC33775.1 KK852556 KDR21433.1 KQ982450 KYQ56223.1 GEZM01015647 GEZM01015646 JAV91494.1 PNF36909.1 GBRD01016953 GDHC01003390 JAG48874.1 JAQ15239.1 GBXI01012040 GBXI01009573 JAD02252.1 JAD04719.1 GALA01001251 JAA93601.1 EAT41829.1 JXJN01022846 KA647017 AFP61646.1 PNF36908.1 KK107024 QOIP01000004 EZA62297.1 RLU24058.1 GECL01002565 JAP03559.1 KQ414620 KOC67681.1 GDHF01024541 JAI27773.1 GAKP01015902 JAC43050.1 PNF36907.1 JR036722 JR036724 AEY57349.1 AEY57351.1 GEZM01015645 JAV91495.1 JR036721 JR036723 AEY57348.1 AEY57350.1 AAAB01008960 GEZM01015644 JAV91496.1 GBXI01000062 JAD14230.1 CH940648 EDW61159.2 GANO01000863 JAB59008.1 GBXI01009274 GBXI01005620 JAD05018.1 JAD08672.1 UFQS01000224 UFQT01000224 SSX01628.1 SSX22008.1 GAMC01016417 JAB90138.1 ATLV01024183 KE525350 KFB50946.1 CH902619 EDV36261.1 AAZO01006743 DS235860 EEB19132.1 CH963850 EDW74773.1 GGFM01000321 MBW21072.1 GGFL01004262 MBW68440.1 GGMS01006323 MBY75526.1 GAKP01015903 JAC43049.1 KRF79821.1 CM000158 EDW92427.1 GGFK01000628 MBW33949.1 GAMC01016419 JAB90136.1 CH933808 EDW09807.1 M62892 AE013599 AY058465 X17472 GAMC01016418 JAB90137.1 CH954179 EDV56898.1 GGFK01000631 MBW33952.1 KRF79820.1 CM002911 KMY96172.1 CH480824 EDW57042.1 GFDL01006802 JAV28243.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000005204
UP000007151
UP000007266
+ More
UP000002358 UP000076502 UP000092444 UP000235965 UP000078492 UP000078541 UP000078540 UP000005205 UP000078542 UP000008820 UP000037069 UP000027135 UP000075809 UP000192223 UP000005203 UP000092443 UP000092460 UP000091820 UP000092445 UP000053097 UP000279307 UP000053825 UP000007062 UP000078200 UP000008792 UP000030765 UP000095300 UP000007801 UP000009046 UP000007798 UP000002282 UP000009192 UP000000803 UP000008711 UP000001292 UP000076408 UP000192221
UP000002358 UP000076502 UP000092444 UP000235965 UP000078492 UP000078541 UP000078540 UP000005205 UP000078542 UP000008820 UP000037069 UP000027135 UP000075809 UP000192223 UP000005203 UP000092443 UP000092460 UP000091820 UP000092445 UP000053097 UP000279307 UP000053825 UP000007062 UP000078200 UP000008792 UP000030765 UP000095300 UP000007801 UP000009046 UP000007798 UP000002282 UP000009192 UP000000803 UP000008711 UP000001292 UP000076408 UP000192221
Interpro
IPR036412
HAD-like_sf
+ More
IPR023214 HAD_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR030332 SERCA1/2
IPR005782 P-type_ATPase_IIA
IPR004014 ATPase_P-typ_cation-transptr_N
IPR018303 ATPase_P-typ_P_site
IPR023214 HAD_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR030332 SERCA1/2
IPR005782 P-type_ATPase_IIA
IPR004014 ATPase_P-typ_cation-transptr_N
IPR018303 ATPase_P-typ_P_site
Gene 3D
ProteinModelPortal
C7AQP4
A0A2A4KA69
A0A3S2NIS6
A0A194QLJ6
O96696
A0A2A4KB22
+ More
A0A2H1W1G5 A0A1L8DF78 H9JAQ8 A0A212EY66 A0A0C9QYZ2 A0A0C9R823 A0A1L8DF68 A0A1B6JB05 A0A1B6K2B5 A0A385MDB2 A0A1B6E9S2 A0A224X6B8 D6WZH8 K7IPT6 A0A1B6CRW6 A0A154PK50 A0A1B6EA81 A0A069DXH6 A0A1B0G7F1 A0A2J7R7V8 A0A151IYX4 A0A023F5T4 A0A195F7F3 A0A195ATM9 A0A158P1I0 A0A195CU94 A0A0P4VR76 A0A1S4FE53 Q175R5 Q175R4 A0A0L0CN78 A0A067REG0 A0A151X776 A0A1Y1N0V8 A0A1W4X7R1 A0A2J7R7U2 A0A0K8S6S5 A0A088ACZ9 A0A0A1X082 T1DHY9 Q175R3 A0A1A9XZK2 A0A1B0BYV5 A0A1A9W106 T1PE27 A0A2J7R7U7 A0A1A9ZUZ6 A0A026X1Y8 A0A0V0G6F0 A0A1W4XH71 A0A0L7RAA5 A0A0K8UMR7 A0A034VML4 A0A2J7R7U1 V9IA48 A0A1Y1N483 V9I895 Q7PPA5-3 A0A1A9VNL8 A0A1Y1N586 A0A1W4X7X2 Q7PPA5-2 A0A0A1XTC4 B4LNZ0 U5EVU1 A0A0A1X2W8 A0A336KD28 W8AZ56 A0A084WL52 A0A1I8QEW5 B3MCM7 E0W0H6 Q7PPA5 A0A1I8QEQ1 B4MRN4 A0A2M3YXR9 A0A2M4CSZ5 A0A2S2QCR1 A0A034VKP1 A0A0Q9W4D5 B4PAA1 A0A2M3ZZM7 W8BAL8 B4KMB9 P22700-2 W8AV60 B3NQ25 A0A2M3ZZP9 A0A0Q9W432 A0A0J9RK49 B4I8W7 A0A182YKV8 A0A1W4V0L4 A0A1Q3FL77
A0A2H1W1G5 A0A1L8DF78 H9JAQ8 A0A212EY66 A0A0C9QYZ2 A0A0C9R823 A0A1L8DF68 A0A1B6JB05 A0A1B6K2B5 A0A385MDB2 A0A1B6E9S2 A0A224X6B8 D6WZH8 K7IPT6 A0A1B6CRW6 A0A154PK50 A0A1B6EA81 A0A069DXH6 A0A1B0G7F1 A0A2J7R7V8 A0A151IYX4 A0A023F5T4 A0A195F7F3 A0A195ATM9 A0A158P1I0 A0A195CU94 A0A0P4VR76 A0A1S4FE53 Q175R5 Q175R4 A0A0L0CN78 A0A067REG0 A0A151X776 A0A1Y1N0V8 A0A1W4X7R1 A0A2J7R7U2 A0A0K8S6S5 A0A088ACZ9 A0A0A1X082 T1DHY9 Q175R3 A0A1A9XZK2 A0A1B0BYV5 A0A1A9W106 T1PE27 A0A2J7R7U7 A0A1A9ZUZ6 A0A026X1Y8 A0A0V0G6F0 A0A1W4XH71 A0A0L7RAA5 A0A0K8UMR7 A0A034VML4 A0A2J7R7U1 V9IA48 A0A1Y1N483 V9I895 Q7PPA5-3 A0A1A9VNL8 A0A1Y1N586 A0A1W4X7X2 Q7PPA5-2 A0A0A1XTC4 B4LNZ0 U5EVU1 A0A0A1X2W8 A0A336KD28 W8AZ56 A0A084WL52 A0A1I8QEW5 B3MCM7 E0W0H6 Q7PPA5 A0A1I8QEQ1 B4MRN4 A0A2M3YXR9 A0A2M4CSZ5 A0A2S2QCR1 A0A034VKP1 A0A0Q9W4D5 B4PAA1 A0A2M3ZZM7 W8BAL8 B4KMB9 P22700-2 W8AV60 B3NQ25 A0A2M3ZZP9 A0A0Q9W432 A0A0J9RK49 B4I8W7 A0A182YKV8 A0A1W4V0L4 A0A1Q3FL77
PDB
6HXB
E-value=0,
Score=3709
Ontologies
GO
GO:0005388
GO:0016021
GO:0033017
GO:0005524
GO:0006816
GO:0070588
GO:0008553
GO:0006874
GO:0046872
GO:0007274
GO:0060047
GO:1903515
GO:0048863
GO:0005789
GO:0030322
GO:1901896
GO:0012505
GO:0006635
GO:1990845
GO:0005783
GO:0007629
GO:0030707
GO:0036335
GO:0051282
GO:0010884
GO:0008610
GO:0016529
GO:0000166
GO:0006974
GO:0001664
GO:0005525
GO:0007264
GO:0003824
PANTHER
Topology
Subcellular location
Membrane
Endoplasmic reticulum membrane
Sarcoplasmic reticulum membrane
Endoplasmic reticulum membrane
Sarcoplasmic reticulum membrane
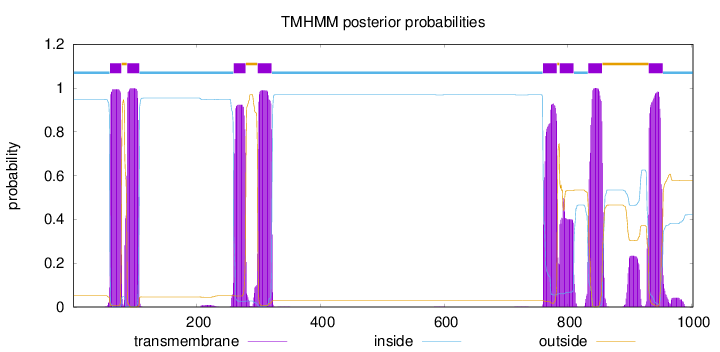
Length:
1002
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
161.22699
Exp number, first 60 AAs:
0.93101
Total prob of N-in:
0.94841
inside
1 - 59
TMhelix
60 - 78
outside
79 - 87
TMhelix
88 - 107
inside
108 - 259
TMhelix
260 - 279
outside
280 - 298
TMhelix
299 - 321
inside
322 - 759
TMhelix
760 - 782
outside
783 - 786
TMhelix
787 - 809
inside
810 - 832
TMhelix
833 - 855
outside
856 - 930
TMhelix
931 - 953
inside
954 - 1002
Population Genetic Test Statistics
Pi
170.961071
Theta
179.042591
Tajima's D
-0.57074
CLR
11.509951
CSRT
0.226038698065097
Interpretation
Uncertain
