Gene
KWMTBOMO05537
Pre Gene Modal
BGIBMGA006605
Annotation
PREDICTED:_UPF0545_protein_C22orf39_homolog_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.138
Sequence
CDS
ATGACAGACAATAATAGGAGAAATGAAAAAAATGAATCGGAAATCCATAAAGAGTCTATTAATACATCCAATACTACAGAAAGTCAATTAGAGGATAAGTGGATGATAAGGAATTGTGACATATATGAAGATGAATATGACGAATGCACAAGTTTCAGAGCTAGATTCCATCAGTACTTCATATTTGGTGAGACATTAGATTGTAACCAATGGAAAAAAGATTATGATAATTGTTGTAAATGGGAATCGAAAAAAGATGTTAAAGCTGCTGAAGCACTTATAAATAGCGAAAAAACAAGACGACTAGAAAGATTAAAAGGACACTACAACAACAGCATATGGAAGAAACGCAGTGCACCACCAGCTAACTGGGATAAGCCATTACCAGAATGGATGTCAAAAAGAGATGAGAACACATACCTTGCGTTAAAAGCTAAAGAATTCAGAGAAGGAAACGTGAAAGAAGAAAAAAGTTTTTGTAGCATTATGTGA
Protein
MTDNNRRNEKNESEIHKESINTSNTTESQLEDKWMIRNCDIYEDEYDECTSFRARFHQYFIFGETLDCNQWKKDYDNCCKWESKKDVKAAEALINSEKTRRLERLKGHYNNSIWKKRSAPPANWDKPLPEWMSKRDENTYLALKAKEFREGNVKEEKSFCSIM
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the aldehyde dehydrogenase family.
Belongs to the PP2C family.
Belongs to the PP2C family.
Uniprot
H9JAR0
A0A2A4K9Y6
A0A194PG35
A0A194QL07
S4PT01
A0A2H1W1R1
+ More
A0A1E1WCH5 A0A212EY64 A0A3S2NZM8 A0A0L7L5F8 W8AQX5 A0A1W4W3T2 A0A1I8M0Q2 A0A0L0C2T2 A0A1B0BR54 A0A1A9Y6B2 A0A1B0FKG7 A0A1I8Q5X7 A0A1A9VX74 A0A1A9ZP69 A0A0C9RC14 A0A034WPJ6 A0A0J7K8I9 B3MJ39 D6X2H5 A0A067QUI1 A0A195DMK9 A0A0K8W9M4 A0A158P1Y7 A0A0K8TR64 A0A1A9WK27 A0A195FKD5 A0A087ZQ23 A0A084VIE1 A0A182IPG0 A0A0M4EFL3 A0A1B6F593 B4KNJ4 A0A182FUS6 A0A2M3ZCX1 A0A1Y1NCH6 B4NMH7 Q293A2 E2AHG5 A0A1B6I5V6 A0A182HGX3 A0A2M4B023 B4GC66 A0A2M4AZV2 A0NGT8 A0A151IKI5 A0A2M4C1Y5 B4LJZ9 A0A182M142 B4J6I5 T1DNJ3 B3NB44 A0A182PC94 A0A182TWB6 A0A182VBR9 A0A2M4C239 A0A3B0J0H3 A0A2M4C1Z7 A0A2A3EG49 A0A1W4WED3 A0A1B0CKZ6 A0A170XTF1 A0A1L8DD00 U5EGX4 A0A195AW93 B4QDA7 B4HPW6 A0A336K5Q7 A0A0P4WPV0 W5JRI0 A0A069DP87 A0A151WT37 A0A0V0G9J7 A0A182YJC8 Q177D2 A0A2J7RBD3 A0A1W4V1K1 A0A023EG02 A0A026WAP9 A0A232ERP5 A0A2S2QKF1 A0A182WEI8 K7IXD8 B0W338 U4UJA7 A0A182N2G6 A0A224Y0S6 N6TCE0 A0A023G732 G3MSG2 A0A1Q3FAD6 Q4QPU3 A0A1J1ISM5 A0A2H8TWT4 A0A023G132
A0A1E1WCH5 A0A212EY64 A0A3S2NZM8 A0A0L7L5F8 W8AQX5 A0A1W4W3T2 A0A1I8M0Q2 A0A0L0C2T2 A0A1B0BR54 A0A1A9Y6B2 A0A1B0FKG7 A0A1I8Q5X7 A0A1A9VX74 A0A1A9ZP69 A0A0C9RC14 A0A034WPJ6 A0A0J7K8I9 B3MJ39 D6X2H5 A0A067QUI1 A0A195DMK9 A0A0K8W9M4 A0A158P1Y7 A0A0K8TR64 A0A1A9WK27 A0A195FKD5 A0A087ZQ23 A0A084VIE1 A0A182IPG0 A0A0M4EFL3 A0A1B6F593 B4KNJ4 A0A182FUS6 A0A2M3ZCX1 A0A1Y1NCH6 B4NMH7 Q293A2 E2AHG5 A0A1B6I5V6 A0A182HGX3 A0A2M4B023 B4GC66 A0A2M4AZV2 A0NGT8 A0A151IKI5 A0A2M4C1Y5 B4LJZ9 A0A182M142 B4J6I5 T1DNJ3 B3NB44 A0A182PC94 A0A182TWB6 A0A182VBR9 A0A2M4C239 A0A3B0J0H3 A0A2M4C1Z7 A0A2A3EG49 A0A1W4WED3 A0A1B0CKZ6 A0A170XTF1 A0A1L8DD00 U5EGX4 A0A195AW93 B4QDA7 B4HPW6 A0A336K5Q7 A0A0P4WPV0 W5JRI0 A0A069DP87 A0A151WT37 A0A0V0G9J7 A0A182YJC8 Q177D2 A0A2J7RBD3 A0A1W4V1K1 A0A023EG02 A0A026WAP9 A0A232ERP5 A0A2S2QKF1 A0A182WEI8 K7IXD8 B0W338 U4UJA7 A0A182N2G6 A0A224Y0S6 N6TCE0 A0A023G732 G3MSG2 A0A1Q3FAD6 Q4QPU3 A0A1J1ISM5 A0A2H8TWT4 A0A023G132
Pubmed
19121390
26354079
23622113
22118469
26227816
24495485
+ More
25315136 26108605 25348373 17994087 18362917 19820115 24845553 21347285 26369729 24438588 28004739 15632085 20798317 12364791 22936249 20920257 23761445 26334808 25244985 17510324 24945155 26483478 24508170 30249741 28648823 20075255 23537049 22216098 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
25315136 26108605 25348373 17994087 18362917 19820115 24845553 21347285 26369729 24438588 28004739 15632085 20798317 12364791 22936249 20920257 23761445 26334808 25244985 17510324 24945155 26483478 24508170 30249741 28648823 20075255 23537049 22216098 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01008558
NWSH01000001
PCG81105.1
KQ459606
KPI91664.1
KQ461198
+ More
KPJ06258.1 GAIX01012898 JAA79662.1 ODYU01005735 SOQ46886.1 GDQN01006376 JAT84678.1 AGBW02011611 OWR46436.1 RSAL01000081 RVE48595.1 JTDY01002870 KOB70566.1 GAMC01019628 GAMC01019622 GAMC01019618 GAMC01019612 JAB86933.1 JRES01000975 KNC26571.1 JXJN01018908 CCAG010009561 GBYB01005790 JAG75557.1 GAKP01001431 JAC57521.1 LBMM01011578 KMQ86753.1 CH902619 EDV38133.1 KQ971372 EFA09858.1 KK853247 KDR09384.1 KQ980724 KYN14130.1 GDHF01004421 JAI47893.1 ADTU01006915 GDAI01000985 JAI16618.1 KQ981523 KYN40459.1 ATLV01013353 KE524854 KFB37735.1 CP012524 ALC42164.1 GECZ01024409 JAS45360.1 CH933808 EDW08953.1 GGFM01005645 MBW26396.1 GEZM01011966 JAV93327.1 CH964282 EDW85566.1 CM000071 EAL24709.1 GL439539 EFN67120.1 GECU01025421 JAS82285.1 APCN01002367 GGFK01013001 MBW46322.1 CH479181 EDW31384.1 GGFK01013002 MBW46323.1 AAAB01008986 EAU75885.2 KQ977199 KYN04986.1 GGFJ01010195 MBW59336.1 CH940648 EDW61653.1 AXCM01001816 CH916367 EDW01986.1 GAMD01002815 JAA98775.1 CH954177 EDV59809.1 GGFJ01010259 MBW59400.1 OUUW01000001 SPP74175.1 GGFJ01010196 MBW59337.1 KZ288254 PBC30717.1 AJWK01016803 GEMB01004058 JAR99203.1 GFDF01009748 JAV04336.1 GANO01003261 JAB56610.1 KQ976731 KYM76305.1 CM000362 CM002911 EDX05894.1 KMY91760.1 CH480816 EDW46635.1 UFQS01000091 UFQT01000091 SSW99182.1 SSX19564.1 GDRN01072735 JAI63510.1 ADMH02000317 ETN67002.1 GBGD01003393 JAC85496.1 KQ982763 KYQ51014.1 GECL01001539 JAP04585.1 CH477378 EAT42260.1 NEVH01005906 PNF38147.1 JXUM01048844 GAPW01005326 KQ561579 JAC08272.1 KXJ78108.1 KK107293 QOIP01000005 EZA53162.1 RLU22966.1 NNAY01002581 OXU21002.1 GGMS01009014 MBY78217.1 AAZX01000012 DS231830 EDS30746.1 KB632328 ERL92528.1 GFTR01002397 JAW14029.1 APGK01043279 KB741014 ENN75408.1 GBBM01005811 JAC29607.1 JO844813 AEO36430.1 GFDL01010501 JAV24544.1 AE013599 BT023673 KX532067 AAF57371.1 AAY85073.1 ANY27877.1 CVRI01000055 CRL01529.1 GFXV01006574 MBW18379.1 GBBL01000720 JAC26600.1
KPJ06258.1 GAIX01012898 JAA79662.1 ODYU01005735 SOQ46886.1 GDQN01006376 JAT84678.1 AGBW02011611 OWR46436.1 RSAL01000081 RVE48595.1 JTDY01002870 KOB70566.1 GAMC01019628 GAMC01019622 GAMC01019618 GAMC01019612 JAB86933.1 JRES01000975 KNC26571.1 JXJN01018908 CCAG010009561 GBYB01005790 JAG75557.1 GAKP01001431 JAC57521.1 LBMM01011578 KMQ86753.1 CH902619 EDV38133.1 KQ971372 EFA09858.1 KK853247 KDR09384.1 KQ980724 KYN14130.1 GDHF01004421 JAI47893.1 ADTU01006915 GDAI01000985 JAI16618.1 KQ981523 KYN40459.1 ATLV01013353 KE524854 KFB37735.1 CP012524 ALC42164.1 GECZ01024409 JAS45360.1 CH933808 EDW08953.1 GGFM01005645 MBW26396.1 GEZM01011966 JAV93327.1 CH964282 EDW85566.1 CM000071 EAL24709.1 GL439539 EFN67120.1 GECU01025421 JAS82285.1 APCN01002367 GGFK01013001 MBW46322.1 CH479181 EDW31384.1 GGFK01013002 MBW46323.1 AAAB01008986 EAU75885.2 KQ977199 KYN04986.1 GGFJ01010195 MBW59336.1 CH940648 EDW61653.1 AXCM01001816 CH916367 EDW01986.1 GAMD01002815 JAA98775.1 CH954177 EDV59809.1 GGFJ01010259 MBW59400.1 OUUW01000001 SPP74175.1 GGFJ01010196 MBW59337.1 KZ288254 PBC30717.1 AJWK01016803 GEMB01004058 JAR99203.1 GFDF01009748 JAV04336.1 GANO01003261 JAB56610.1 KQ976731 KYM76305.1 CM000362 CM002911 EDX05894.1 KMY91760.1 CH480816 EDW46635.1 UFQS01000091 UFQT01000091 SSW99182.1 SSX19564.1 GDRN01072735 JAI63510.1 ADMH02000317 ETN67002.1 GBGD01003393 JAC85496.1 KQ982763 KYQ51014.1 GECL01001539 JAP04585.1 CH477378 EAT42260.1 NEVH01005906 PNF38147.1 JXUM01048844 GAPW01005326 KQ561579 JAC08272.1 KXJ78108.1 KK107293 QOIP01000005 EZA53162.1 RLU22966.1 NNAY01002581 OXU21002.1 GGMS01009014 MBY78217.1 AAZX01000012 DS231830 EDS30746.1 KB632328 ERL92528.1 GFTR01002397 JAW14029.1 APGK01043279 KB741014 ENN75408.1 GBBM01005811 JAC29607.1 JO844813 AEO36430.1 GFDL01010501 JAV24544.1 AE013599 BT023673 KX532067 AAF57371.1 AAY85073.1 ANY27877.1 CVRI01000055 CRL01529.1 GFXV01006574 MBW18379.1 GBBL01000720 JAC26600.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000192223 UP000095301 UP000037069 UP000092460 UP000092443 UP000092444 UP000095300 UP000078200 UP000092445 UP000036403 UP000007801 UP000007266 UP000027135 UP000078492 UP000005205 UP000091820 UP000078541 UP000005203 UP000030765 UP000075880 UP000092553 UP000009192 UP000069272 UP000007798 UP000001819 UP000000311 UP000075840 UP000008744 UP000007062 UP000078542 UP000008792 UP000075883 UP000001070 UP000008711 UP000075885 UP000075902 UP000075903 UP000268350 UP000242457 UP000092461 UP000078540 UP000000304 UP000001292 UP000000673 UP000075809 UP000076408 UP000008820 UP000235965 UP000192221 UP000069940 UP000249989 UP000053097 UP000279307 UP000215335 UP000075920 UP000002358 UP000002320 UP000030742 UP000075884 UP000019118 UP000000803 UP000183832
UP000037510 UP000192223 UP000095301 UP000037069 UP000092460 UP000092443 UP000092444 UP000095300 UP000078200 UP000092445 UP000036403 UP000007801 UP000007266 UP000027135 UP000078492 UP000005205 UP000091820 UP000078541 UP000005203 UP000030765 UP000075880 UP000092553 UP000009192 UP000069272 UP000007798 UP000001819 UP000000311 UP000075840 UP000008744 UP000007062 UP000078542 UP000008792 UP000075883 UP000001070 UP000008711 UP000075885 UP000075902 UP000075903 UP000268350 UP000242457 UP000092461 UP000078540 UP000000304 UP000001292 UP000000673 UP000075809 UP000076408 UP000008820 UP000235965 UP000192221 UP000069940 UP000249989 UP000053097 UP000279307 UP000215335 UP000075920 UP000002358 UP000002320 UP000030742 UP000075884 UP000019118 UP000000803 UP000183832
Interpro
IPR021475
DUF3128
+ More
IPR016161 Ald_DH/histidinol_DH
IPR016163 Ald_DH_C
IPR016162 Ald_DH_N
IPR029510 Ald_DH_CS_GLU
IPR015590 Aldehyde_DH_dom
IPR015655 PP2C
IPR036457 PPM-type_dom_sf
IPR000222 PP2C_BS
IPR001932 PPM-type_phosphatase_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR012934 Znf_AD
IPR016161 Ald_DH/histidinol_DH
IPR016163 Ald_DH_C
IPR016162 Ald_DH_N
IPR029510 Ald_DH_CS_GLU
IPR015590 Aldehyde_DH_dom
IPR015655 PP2C
IPR036457 PPM-type_dom_sf
IPR000222 PP2C_BS
IPR001932 PPM-type_phosphatase_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR012934 Znf_AD
Gene 3D
CDD
ProteinModelPortal
H9JAR0
A0A2A4K9Y6
A0A194PG35
A0A194QL07
S4PT01
A0A2H1W1R1
+ More
A0A1E1WCH5 A0A212EY64 A0A3S2NZM8 A0A0L7L5F8 W8AQX5 A0A1W4W3T2 A0A1I8M0Q2 A0A0L0C2T2 A0A1B0BR54 A0A1A9Y6B2 A0A1B0FKG7 A0A1I8Q5X7 A0A1A9VX74 A0A1A9ZP69 A0A0C9RC14 A0A034WPJ6 A0A0J7K8I9 B3MJ39 D6X2H5 A0A067QUI1 A0A195DMK9 A0A0K8W9M4 A0A158P1Y7 A0A0K8TR64 A0A1A9WK27 A0A195FKD5 A0A087ZQ23 A0A084VIE1 A0A182IPG0 A0A0M4EFL3 A0A1B6F593 B4KNJ4 A0A182FUS6 A0A2M3ZCX1 A0A1Y1NCH6 B4NMH7 Q293A2 E2AHG5 A0A1B6I5V6 A0A182HGX3 A0A2M4B023 B4GC66 A0A2M4AZV2 A0NGT8 A0A151IKI5 A0A2M4C1Y5 B4LJZ9 A0A182M142 B4J6I5 T1DNJ3 B3NB44 A0A182PC94 A0A182TWB6 A0A182VBR9 A0A2M4C239 A0A3B0J0H3 A0A2M4C1Z7 A0A2A3EG49 A0A1W4WED3 A0A1B0CKZ6 A0A170XTF1 A0A1L8DD00 U5EGX4 A0A195AW93 B4QDA7 B4HPW6 A0A336K5Q7 A0A0P4WPV0 W5JRI0 A0A069DP87 A0A151WT37 A0A0V0G9J7 A0A182YJC8 Q177D2 A0A2J7RBD3 A0A1W4V1K1 A0A023EG02 A0A026WAP9 A0A232ERP5 A0A2S2QKF1 A0A182WEI8 K7IXD8 B0W338 U4UJA7 A0A182N2G6 A0A224Y0S6 N6TCE0 A0A023G732 G3MSG2 A0A1Q3FAD6 Q4QPU3 A0A1J1ISM5 A0A2H8TWT4 A0A023G132
A0A1E1WCH5 A0A212EY64 A0A3S2NZM8 A0A0L7L5F8 W8AQX5 A0A1W4W3T2 A0A1I8M0Q2 A0A0L0C2T2 A0A1B0BR54 A0A1A9Y6B2 A0A1B0FKG7 A0A1I8Q5X7 A0A1A9VX74 A0A1A9ZP69 A0A0C9RC14 A0A034WPJ6 A0A0J7K8I9 B3MJ39 D6X2H5 A0A067QUI1 A0A195DMK9 A0A0K8W9M4 A0A158P1Y7 A0A0K8TR64 A0A1A9WK27 A0A195FKD5 A0A087ZQ23 A0A084VIE1 A0A182IPG0 A0A0M4EFL3 A0A1B6F593 B4KNJ4 A0A182FUS6 A0A2M3ZCX1 A0A1Y1NCH6 B4NMH7 Q293A2 E2AHG5 A0A1B6I5V6 A0A182HGX3 A0A2M4B023 B4GC66 A0A2M4AZV2 A0NGT8 A0A151IKI5 A0A2M4C1Y5 B4LJZ9 A0A182M142 B4J6I5 T1DNJ3 B3NB44 A0A182PC94 A0A182TWB6 A0A182VBR9 A0A2M4C239 A0A3B0J0H3 A0A2M4C1Z7 A0A2A3EG49 A0A1W4WED3 A0A1B0CKZ6 A0A170XTF1 A0A1L8DD00 U5EGX4 A0A195AW93 B4QDA7 B4HPW6 A0A336K5Q7 A0A0P4WPV0 W5JRI0 A0A069DP87 A0A151WT37 A0A0V0G9J7 A0A182YJC8 Q177D2 A0A2J7RBD3 A0A1W4V1K1 A0A023EG02 A0A026WAP9 A0A232ERP5 A0A2S2QKF1 A0A182WEI8 K7IXD8 B0W338 U4UJA7 A0A182N2G6 A0A224Y0S6 N6TCE0 A0A023G732 G3MSG2 A0A1Q3FAD6 Q4QPU3 A0A1J1ISM5 A0A2H8TWT4 A0A023G132
Ontologies
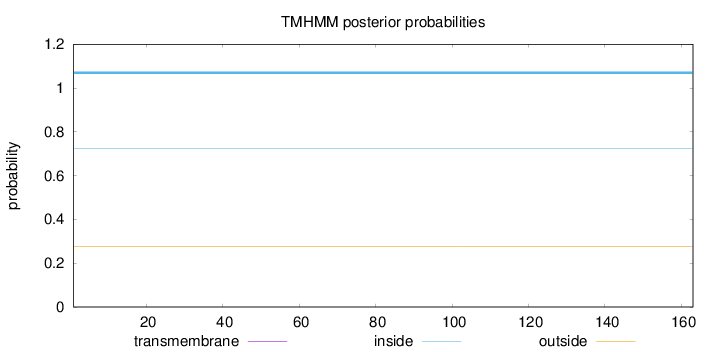
Topology
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.72227
inside
1 - 163
Population Genetic Test Statistics
Pi
224.266038
Theta
214.644966
Tajima's D
0.409064
CLR
0
CSRT
0.494675266236688
Interpretation
Uncertain
