Gene
KWMTBOMO05527
Annotation
PREDICTED:_probable_tRNA_(uracil-O(2)-)-methyltransferase_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 1.881
Sequence
CDS
ATGATTGTCTTGAATGCACACTGTGATGATGAAAACTCAAAATCATACATGTGGCTCAAGAATAAGCTCTTACCACAGTTTATCAAATGGACAACTGAAACTGAAAGCAATAATGGAAAGAAGAAAATCTGCACAGCGTCTTTAACTTTAGTATCAAGTAGTAAATATTTTGAAAAATATAATGAACTAAAACTCAAGTATGGGAAAGACTTAGTTAAGATTTGGCCAGAATGTACTGATCCTACAAAATTTGTATATGAAGATGTTGCTATTGCAACTTATTTGCTCCTGCTTTGGGAAGACCGAAGTTTAGTAAAGAAACAAACATTTGTGGATCTTGGATGTGGCAATGGTTTATTGGTGTATATATTATGTAAAGAGGGCCATGCAGGACTTGGAATTGATGTAAGAAAAAGAGAAATTTGGGATATGTATCCGCCTGAGGTAAAATTGAAGATGAAAACTATTGTACCCTCAGAAAGCAATCTGTTTCCAAATGCTGATTGGATTATTGGTAACCATTCTGATGAATTGACACCTTGGATTCCTGTCATAGCAGCTAAGAGTTCTTACAAATGCAACTTTTTCCTATTGCCATGTTGTGCATTCAACTTTGATGGGTCCAAATACCAACGAGTGGATTCTAAAAAAAGTCAATACACTGAATATTTAGAACACGTAAAGAAAATTTGTGAAGATTGTGGTTTTATAACTGATCTTGACAGACTTAAAATACCAAGTACGAAGAGAATATGTTTAGTAAGCAATGGTCGAATGTACAGTCCAGACACTTATAAAGACAGTATTAATAAAATTTCTAAAATTTTCAAAGAGAAACATGCACGTGGTAATGTAGAAAATGACACCTGGTTGGCAGATTTTAAAGCTAGAGAATCTACACAGAAAGTACGGAATTGTACACAGTTGGATAAAAACCTAATAGAGTCAATAGTAAAAATAGTGACAGACTGTTTGCTTGAAGGGTGCAGTAAAGACTGTAATGAACAATGGTCTGTTGGAAAAATTGTTGAAATAAGTGAACTAGTTTCACTAATTCCAAAGCAAAATCTTATTAAACTTAAATCTGAATGTGGGGGTTTGCAAACTTTATTAAAGAACAATCATAACATATTTCTTGTTTCTGGTGGTAAAGTGCAGTTGAGGTATCCTAAAACAGTAGACCAAGTGATTACTATTCAAAAGAGACAAAAAATAATTGACACAAAAATACAAGTAAAGCCTTGCTGGTTCCACAACAACCATCCTCAAGGGTGTCCCCTGTCTAGCATCAATTGTAGCTTCCTACACAGTAAAGGATAA
Protein
MIVLNAHCDDENSKSYMWLKNKLLPQFIKWTTETESNNGKKKICTASLTLVSSSKYFEKYNELKLKYGKDLVKIWPECTDPTKFVYEDVAIATYLLLLWEDRSLVKKQTFVDLGCGNGLLVYILCKEGHAGLGIDVRKREIWDMYPPEVKLKMKTIVPSESNLFPNADWIIGNHSDELTPWIPVIAAKSSYKCNFFLLPCCAFNFDGSKYQRVDSKKSQYTEYLEHVKKICEDCGFITDLDRLKIPSTKRICLVSNGRMYSPDTYKDSINKISKIFKEKHARGNVENDTWLADFKARESTQKVRNCTQLDKNLIESIVKIVTDCLLEGCSKDCNEQWSVGKIVEISELVSLIPKQNLIKLKSECGGLQTLLKNNHNIFLVSGGKVQLRYPKTVDQVITIQKRQKIIDTKIQVKPCWFHNNHPQGCPLSSINCSFLHSKG
Summary
Similarity
Belongs to the complex I LYR family.
Uniprot
A0A2A4KBC0
A0A2H1W1D3
A0A0L7L501
A0A3S2NIR1
A0A212EY67
A0A194PE88
+ More
A0A194QMS2 A0A158NNA7 E2B3H2 E9IUF5 A0A195AV02 A0A151XA85 A0A151IT28 A0A195CS06 F4WLH8 A0A026WVH7 A0A0J7L5X8 K7IN52 A0A232F846 W5JB33 A0A182G8G1 A0A0T6BE59 A0A182FDT0 A0A182TD64 A0A182JYY8 Q7Q535 E2AJ27 A0A067RAV4 A0A2J7QDP6 A0A182I4P3 A0A087ZWJ4 A0A084WK01 A0A182RJN8 A0A182UQM1 A0A182X4P6 A0A182PMH5 A0A310SIE1 A0A2A3EL51 V9IID9 A0A182LX36 A0A1B6GY60 Q16HA1 A0A182N977 A0A182QM27 A0A182IU91 A0A154P0R0 A0A182LK68 A0A182WBK6 A0A195FWB2 A0A3P9HGH9 A0A3P9KMX4 A0A182Y2E5 A0A3S2NZL0 H2LNP4 A0A0L7QVF0 A0A3P8UL25 A0A3B3DDY9 A0A1B6MUQ7 A0A3B5B769 A0A3Q1GDU6 A0A1B6DAG1 A0A2U9B173 A0A3Q3L7M4 A0A3P9J1K3 I3J089 A0A1S3N6X9 A0A3B4FEN2 A0A3P8NMZ8 A0A3P9D3K1 W5KVK8 G3Q6Z6 A0A0A9Z0V6 A0A0K8SGC5 A0A3B3IKC0 A0A3Q2WK78 A0A3B4VAH8 A0A3Q1AK00 H2TTI4 A0A3P8T3S3 A0A3Q3JD81 A0A1W4XCV2 A0A3Q3GU02 A0A3Q3QKJ3 A0A3Q3J7I7 A0A0P4VVC2 A0A3Q3J7U2 A0A3B3VFU3 A0A087XL22 A0A3B3WFT0 M4AN05 A0A3P9PG75 A0A3B4XIB4 A0A1A8NX50 A0A3Q2CKX7 E9QD66 A0A3R7PCJ4 A0A0F8BV78 A0A1A8MG19 T1HP40 A0A3P9A416
A0A194QMS2 A0A158NNA7 E2B3H2 E9IUF5 A0A195AV02 A0A151XA85 A0A151IT28 A0A195CS06 F4WLH8 A0A026WVH7 A0A0J7L5X8 K7IN52 A0A232F846 W5JB33 A0A182G8G1 A0A0T6BE59 A0A182FDT0 A0A182TD64 A0A182JYY8 Q7Q535 E2AJ27 A0A067RAV4 A0A2J7QDP6 A0A182I4P3 A0A087ZWJ4 A0A084WK01 A0A182RJN8 A0A182UQM1 A0A182X4P6 A0A182PMH5 A0A310SIE1 A0A2A3EL51 V9IID9 A0A182LX36 A0A1B6GY60 Q16HA1 A0A182N977 A0A182QM27 A0A182IU91 A0A154P0R0 A0A182LK68 A0A182WBK6 A0A195FWB2 A0A3P9HGH9 A0A3P9KMX4 A0A182Y2E5 A0A3S2NZL0 H2LNP4 A0A0L7QVF0 A0A3P8UL25 A0A3B3DDY9 A0A1B6MUQ7 A0A3B5B769 A0A3Q1GDU6 A0A1B6DAG1 A0A2U9B173 A0A3Q3L7M4 A0A3P9J1K3 I3J089 A0A1S3N6X9 A0A3B4FEN2 A0A3P8NMZ8 A0A3P9D3K1 W5KVK8 G3Q6Z6 A0A0A9Z0V6 A0A0K8SGC5 A0A3B3IKC0 A0A3Q2WK78 A0A3B4VAH8 A0A3Q1AK00 H2TTI4 A0A3P8T3S3 A0A3Q3JD81 A0A1W4XCV2 A0A3Q3GU02 A0A3Q3QKJ3 A0A3Q3J7I7 A0A0P4VVC2 A0A3Q3J7U2 A0A3B3VFU3 A0A087XL22 A0A3B3WFT0 M4AN05 A0A3P9PG75 A0A3B4XIB4 A0A1A8NX50 A0A3Q2CKX7 E9QD66 A0A3R7PCJ4 A0A0F8BV78 A0A1A8MG19 T1HP40 A0A3P9A416
Pubmed
26227816
22118469
26354079
21347285
20798317
21282665
+ More
21719571 24508170 30249741 20075255 28648823 20920257 23761445 26483478 12364791 14747013 17210077 24845553 24438588 17510324 20966253 17554307 25244985 24487278 29451363 25186727 25329095 25401762 26823975 21551351 23542700 23594743 25835551 25069045
21719571 24508170 30249741 20075255 28648823 20920257 23761445 26483478 12364791 14747013 17210077 24845553 24438588 17510324 20966253 17554307 25244985 24487278 29451363 25186727 25329095 25401762 26823975 21551351 23542700 23594743 25835551 25069045
EMBL
NWSH01000001
PCG81103.1
ODYU01005735
SOQ46889.1
JTDY01002870
KOB70568.1
+ More
RSAL01000081 RVE48593.1 AGBW02011611 OWR46438.1 KQ459606 KPI91666.1 KQ461198 KPJ06260.1 ADTU01021199 GL445346 EFN89724.1 GL765974 EFZ15784.1 KQ976738 KYM75855.1 KQ982351 KYQ57285.1 KQ981037 KYN10159.1 KQ977372 KYN03277.1 GL888208 EGI64914.1 KK107109 QOIP01000008 EZA59094.1 RLU19886.1 LBMM01000586 KMQ98026.1 NNAY01000764 OXU26629.1 ADMH02001746 ETN61211.1 JXUM01151982 KQ571343 KXJ68193.1 LJIG01001341 KRT85599.1 AAAB01008960 EAA11433.4 GL439932 EFN66559.1 KK852772 KDR16845.1 NEVH01015817 PNF26712.1 APCN01002341 ATLV01024090 KE525349 KFB50545.1 KQ763441 OAD55102.1 KZ288215 PBC32440.1 JR047417 AEY60412.1 AXCM01001604 GECZ01002418 JAS67351.1 CH478188 EAT33623.1 AXCN02000557 KQ434792 KZC05437.1 KQ981208 KYN44733.1 CM012454 RVE60530.1 KQ414727 KOC62600.1 GEBQ01000312 JAT39665.1 GEDC01014630 JAS22668.1 CP026244 AWO97617.1 AERX01055572 AERX01055573 GBHO01004697 GBHO01004696 GDHC01010239 GDHC01000954 GDHC01000952 JAG38907.1 JAG38908.1 JAQ08390.1 JAQ17675.1 JAQ17677.1 GBRD01013996 JAG51830.1 GDRN01093374 JAI59902.1 AYCK01004350 HAEG01005160 SBR73434.1 BX088542 QCYY01002901 ROT66706.1 KQ042189 KKF18254.1 HAEF01014568 SBR55727.1 ACPB03008068
RSAL01000081 RVE48593.1 AGBW02011611 OWR46438.1 KQ459606 KPI91666.1 KQ461198 KPJ06260.1 ADTU01021199 GL445346 EFN89724.1 GL765974 EFZ15784.1 KQ976738 KYM75855.1 KQ982351 KYQ57285.1 KQ981037 KYN10159.1 KQ977372 KYN03277.1 GL888208 EGI64914.1 KK107109 QOIP01000008 EZA59094.1 RLU19886.1 LBMM01000586 KMQ98026.1 NNAY01000764 OXU26629.1 ADMH02001746 ETN61211.1 JXUM01151982 KQ571343 KXJ68193.1 LJIG01001341 KRT85599.1 AAAB01008960 EAA11433.4 GL439932 EFN66559.1 KK852772 KDR16845.1 NEVH01015817 PNF26712.1 APCN01002341 ATLV01024090 KE525349 KFB50545.1 KQ763441 OAD55102.1 KZ288215 PBC32440.1 JR047417 AEY60412.1 AXCM01001604 GECZ01002418 JAS67351.1 CH478188 EAT33623.1 AXCN02000557 KQ434792 KZC05437.1 KQ981208 KYN44733.1 CM012454 RVE60530.1 KQ414727 KOC62600.1 GEBQ01000312 JAT39665.1 GEDC01014630 JAS22668.1 CP026244 AWO97617.1 AERX01055572 AERX01055573 GBHO01004697 GBHO01004696 GDHC01010239 GDHC01000954 GDHC01000952 JAG38907.1 JAG38908.1 JAQ08390.1 JAQ17675.1 JAQ17677.1 GBRD01013996 JAG51830.1 GDRN01093374 JAI59902.1 AYCK01004350 HAEG01005160 SBR73434.1 BX088542 QCYY01002901 ROT66706.1 KQ042189 KKF18254.1 HAEF01014568 SBR55727.1 ACPB03008068
Proteomes
UP000218220
UP000037510
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000005205 UP000008237 UP000078540 UP000075809 UP000078492 UP000078542 UP000007755 UP000053097 UP000279307 UP000036403 UP000002358 UP000215335 UP000000673 UP000069940 UP000249989 UP000069272 UP000075902 UP000075881 UP000007062 UP000000311 UP000027135 UP000235965 UP000075840 UP000005203 UP000030765 UP000075900 UP000075903 UP000076407 UP000075885 UP000242457 UP000075883 UP000008820 UP000075884 UP000075886 UP000075880 UP000076502 UP000075882 UP000075920 UP000078541 UP000265200 UP000265180 UP000076408 UP000001038 UP000053825 UP000265120 UP000261560 UP000261400 UP000257200 UP000246464 UP000261640 UP000005207 UP000087266 UP000261460 UP000265100 UP000265160 UP000018467 UP000007635 UP000264840 UP000261420 UP000257160 UP000005226 UP000265080 UP000261600 UP000192223 UP000261660 UP000261500 UP000028760 UP000261480 UP000002852 UP000242638 UP000261360 UP000265020 UP000000437 UP000283509 UP000015103 UP000265140
UP000005205 UP000008237 UP000078540 UP000075809 UP000078492 UP000078542 UP000007755 UP000053097 UP000279307 UP000036403 UP000002358 UP000215335 UP000000673 UP000069940 UP000249989 UP000069272 UP000075902 UP000075881 UP000007062 UP000000311 UP000027135 UP000235965 UP000075840 UP000005203 UP000030765 UP000075900 UP000075903 UP000076407 UP000075885 UP000242457 UP000075883 UP000008820 UP000075884 UP000075886 UP000075880 UP000076502 UP000075882 UP000075920 UP000078541 UP000265200 UP000265180 UP000076408 UP000001038 UP000053825 UP000265120 UP000261560 UP000261400 UP000257200 UP000246464 UP000261640 UP000005207 UP000087266 UP000261460 UP000265100 UP000265160 UP000018467 UP000007635 UP000264840 UP000261420 UP000257160 UP000005226 UP000265080 UP000261600 UP000192223 UP000261660 UP000261500 UP000028760 UP000261480 UP000002852 UP000242638 UP000261360 UP000265020 UP000000437 UP000283509 UP000015103 UP000265140
Interpro
Gene 3D
ProteinModelPortal
A0A2A4KBC0
A0A2H1W1D3
A0A0L7L501
A0A3S2NIR1
A0A212EY67
A0A194PE88
+ More
A0A194QMS2 A0A158NNA7 E2B3H2 E9IUF5 A0A195AV02 A0A151XA85 A0A151IT28 A0A195CS06 F4WLH8 A0A026WVH7 A0A0J7L5X8 K7IN52 A0A232F846 W5JB33 A0A182G8G1 A0A0T6BE59 A0A182FDT0 A0A182TD64 A0A182JYY8 Q7Q535 E2AJ27 A0A067RAV4 A0A2J7QDP6 A0A182I4P3 A0A087ZWJ4 A0A084WK01 A0A182RJN8 A0A182UQM1 A0A182X4P6 A0A182PMH5 A0A310SIE1 A0A2A3EL51 V9IID9 A0A182LX36 A0A1B6GY60 Q16HA1 A0A182N977 A0A182QM27 A0A182IU91 A0A154P0R0 A0A182LK68 A0A182WBK6 A0A195FWB2 A0A3P9HGH9 A0A3P9KMX4 A0A182Y2E5 A0A3S2NZL0 H2LNP4 A0A0L7QVF0 A0A3P8UL25 A0A3B3DDY9 A0A1B6MUQ7 A0A3B5B769 A0A3Q1GDU6 A0A1B6DAG1 A0A2U9B173 A0A3Q3L7M4 A0A3P9J1K3 I3J089 A0A1S3N6X9 A0A3B4FEN2 A0A3P8NMZ8 A0A3P9D3K1 W5KVK8 G3Q6Z6 A0A0A9Z0V6 A0A0K8SGC5 A0A3B3IKC0 A0A3Q2WK78 A0A3B4VAH8 A0A3Q1AK00 H2TTI4 A0A3P8T3S3 A0A3Q3JD81 A0A1W4XCV2 A0A3Q3GU02 A0A3Q3QKJ3 A0A3Q3J7I7 A0A0P4VVC2 A0A3Q3J7U2 A0A3B3VFU3 A0A087XL22 A0A3B3WFT0 M4AN05 A0A3P9PG75 A0A3B4XIB4 A0A1A8NX50 A0A3Q2CKX7 E9QD66 A0A3R7PCJ4 A0A0F8BV78 A0A1A8MG19 T1HP40 A0A3P9A416
A0A194QMS2 A0A158NNA7 E2B3H2 E9IUF5 A0A195AV02 A0A151XA85 A0A151IT28 A0A195CS06 F4WLH8 A0A026WVH7 A0A0J7L5X8 K7IN52 A0A232F846 W5JB33 A0A182G8G1 A0A0T6BE59 A0A182FDT0 A0A182TD64 A0A182JYY8 Q7Q535 E2AJ27 A0A067RAV4 A0A2J7QDP6 A0A182I4P3 A0A087ZWJ4 A0A084WK01 A0A182RJN8 A0A182UQM1 A0A182X4P6 A0A182PMH5 A0A310SIE1 A0A2A3EL51 V9IID9 A0A182LX36 A0A1B6GY60 Q16HA1 A0A182N977 A0A182QM27 A0A182IU91 A0A154P0R0 A0A182LK68 A0A182WBK6 A0A195FWB2 A0A3P9HGH9 A0A3P9KMX4 A0A182Y2E5 A0A3S2NZL0 H2LNP4 A0A0L7QVF0 A0A3P8UL25 A0A3B3DDY9 A0A1B6MUQ7 A0A3B5B769 A0A3Q1GDU6 A0A1B6DAG1 A0A2U9B173 A0A3Q3L7M4 A0A3P9J1K3 I3J089 A0A1S3N6X9 A0A3B4FEN2 A0A3P8NMZ8 A0A3P9D3K1 W5KVK8 G3Q6Z6 A0A0A9Z0V6 A0A0K8SGC5 A0A3B3IKC0 A0A3Q2WK78 A0A3B4VAH8 A0A3Q1AK00 H2TTI4 A0A3P8T3S3 A0A3Q3JD81 A0A1W4XCV2 A0A3Q3GU02 A0A3Q3QKJ3 A0A3Q3J7I7 A0A0P4VVC2 A0A3Q3J7U2 A0A3B3VFU3 A0A087XL22 A0A3B3WFT0 M4AN05 A0A3P9PG75 A0A3B4XIB4 A0A1A8NX50 A0A3Q2CKX7 E9QD66 A0A3R7PCJ4 A0A0F8BV78 A0A1A8MG19 T1HP40 A0A3P9A416
Ontologies
GO
PANTHER
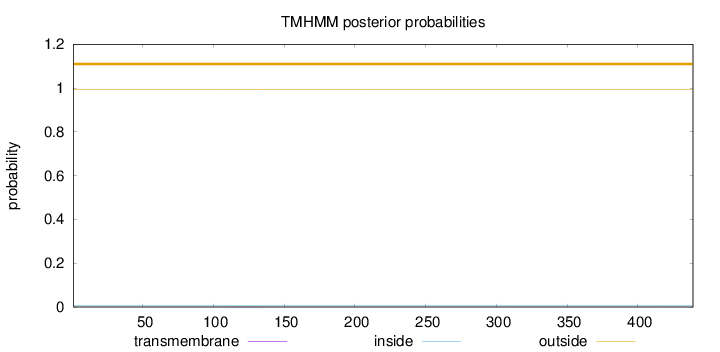
Topology
Length:
439
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01065
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00600
outside
1 - 439
Population Genetic Test Statistics
Pi
0.731819
Theta
46.937774
Tajima's D
-2.385448
CLR
21.068866
CSRT
0.0015999200039998
Interpretation
Possibly Positive selection
