Gene
KWMTBOMO05524 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006733
Annotation
PREDICTED:_fatty_acid-binding_protein_12-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.516
Sequence
CDS
ATGCCGTCAATCGCTGGAAAGTACCAGCACTACAAGAACGAAGACATTGATGATTATTTCAGTGCGGTCGGTGTACCATTTATGGGTCGAAAAATGATGTCAATGTCATCACCATTAATGGAAATTTCAATTGATGGGGATACTATGACTATAAAAAATTCATCATTGTTACGAACAGTAGAACACAAATTTAAGCTTGGCGAAGAATATGAAGAGAAAATGCCCAATTCCATTATAAAAAGTGTCACAACAATCACCAATGATAGTGAAATGGAGACAAAATCTGTTATACCTGACACAGGAGCTAAATGTGGACGTCACTATCAGTTTACTGATGAAGAATGTATTGTGACTCTCACACATGAAAATGCAAGTGTTTCTGGGAAACGCTACTTTCGCAGAGTGCAGTAG
Protein
MPSIAGKYQHYKNEDIDDYFSAVGVPFMGRKMMSMSSPLMEISIDGDTMTIKNSSLLRTVEHKFKLGEEYEEKMPNSIIKSVTTITNDSEMETKSVIPDTGAKCGRHYQFTDEECIVTLTHENASVSGKRYFRRVQ
Summary
Similarity
Belongs to the calycin superfamily. Fatty-acid binding protein (FABP) family.
Uniprot
H9JB38
S4PR71
A0A3S2L9C7
A0A2A4K9X7
A0A2A4KAT4
A0A194QRU9
+ More
A0A1E1WR56 A0A194PG40 A0A2H1W239 A0A212EK27 I4DL17 A0A1E1VZL1 D6X2H6 A0A0N1ITP0 A0A2P8ZJ00 A0A087ZPP1 S4S5K9 A0A232FGE3 K7IQ80 A0A1W4WFN5 A0A2A3EG77 A0A0L7R166 U4UEX6 N6U9W4 A0A1B6EE81 A0A0P5DJW0 A0A0P5T9I6 A0A0P4ZWH9 A0A0P6AP56 A0A1D2N5D4 A0A0P6D1A5 A0A226DEB1 A0A0P5L708 A0A0P5HPX8 A0A0P5IR43 A0A0P5KTU4 A0A0P6H9Q8 A0A0N8A7S3 R7TGQ9 A0A2I9LNZ4 A0A0U2KD12 D6WBV4 A0A0U2UEZ0 E2BED2 B0KZJ6 A0A0U2LEV9 A0A3R7LUW6 V5H4N0 A0A0U2TK48 A0A0U2V2N8 A0A0U2T5Y2 A0A1D1UF68 A0A158P1Y8 A0A1W7RAI2 R4WCK5 A0A0U2T8A5 B0KZK2 A0A0J7KMQ5 A0A162N623 R4WCU0 A7XZL4 Q177Y4 T1E383 Q66RP5 A0A1Y1MPR8 B3P4J3 I3MNX9 A0A336JZJ3
A0A1E1WR56 A0A194PG40 A0A2H1W239 A0A212EK27 I4DL17 A0A1E1VZL1 D6X2H6 A0A0N1ITP0 A0A2P8ZJ00 A0A087ZPP1 S4S5K9 A0A232FGE3 K7IQ80 A0A1W4WFN5 A0A2A3EG77 A0A0L7R166 U4UEX6 N6U9W4 A0A1B6EE81 A0A0P5DJW0 A0A0P5T9I6 A0A0P4ZWH9 A0A0P6AP56 A0A1D2N5D4 A0A0P6D1A5 A0A226DEB1 A0A0P5L708 A0A0P5HPX8 A0A0P5IR43 A0A0P5KTU4 A0A0P6H9Q8 A0A0N8A7S3 R7TGQ9 A0A2I9LNZ4 A0A0U2KD12 D6WBV4 A0A0U2UEZ0 E2BED2 B0KZJ6 A0A0U2LEV9 A0A3R7LUW6 V5H4N0 A0A0U2TK48 A0A0U2V2N8 A0A0U2T5Y2 A0A1D1UF68 A0A158P1Y8 A0A1W7RAI2 R4WCK5 A0A0U2T8A5 B0KZK2 A0A0J7KMQ5 A0A162N623 R4WCU0 A7XZL4 Q177Y4 T1E383 Q66RP5 A0A1Y1MPR8 B3P4J3 I3MNX9 A0A336JZJ3
Pubmed
EMBL
BABH01008537
GAIX01014988
JAA77572.1
RSAL01000081
RVE48590.1
NWSH01000001
+ More
PCG81095.1 PCG81096.1 KQ461198 KPJ06266.1 GDQN01001657 JAT89397.1 KQ459606 KPI91669.1 ODYU01005735 SOQ46892.1 AGBW02014334 OWR41847.1 AK401985 BAM18607.1 GDQN01010887 JAT80167.1 KQ971372 EFA10745.2 KQ435763 KOX75408.1 PYGN01000041 PSN56475.1 HQ828078 HQ828080 AET36540.1 AET36541.1 NNAY01000281 OXU29528.1 KZ288254 PBC30718.1 KQ414668 KOC64588.1 KB632328 ERL92529.1 APGK01043278 APGK01043279 KB741014 ENN75407.1 GEDC01001081 JAS36217.1 GDIP01156349 LRGB01002481 JAJ67053.1 KZS07417.1 GDIP01129164 JAL74550.1 GDIP01211405 JAJ11997.1 GDIP01040173 JAM63542.1 LJIJ01000205 ODN00478.1 GDIP01007204 JAM96511.1 LNIX01000020 OXA43912.1 GDIQ01179334 JAK72391.1 GDIQ01224870 JAK26855.1 GDIQ01234869 JAK16856.1 GDIQ01179333 JAK72392.1 GDIQ01022674 JAN72063.1 GDIP01174153 JAJ49249.1 AMQN01013196 KB310036 ELT92677.1 GFWZ01000120 MBW20110.1 KT754480 ALS04314.1 KQ971315 EEZ99309.1 KT754479 KT754481 ALS04315.1 GL447768 EFN85977.1 DQ979802 ABL09307.1 KT754478 ALS04312.1 QCYY01003397 ROT63697.1 GALX01000698 JAB67768.1 KT754735 ALS04569.1 KT755010 ALS04844.1 KT755063 ALS04897.1 BDGG01000001 GAU88111.1 ADTU01006917 GFAH01000229 JAV48160.1 AK416948 BAN20163.1 KT755062 ALS04896.1 DQ979808 ABL09313.1 LBMM01005278 KMQ91658.1 AZHF01000004 OAA76109.1 AK416989 BAN20204.1 EU106629 ABU97480.1 CH477369 EAT42473.1 GALA01000600 JAA94252.1 AY710432 AAU11502.1 GEZM01024990 JAV87652.1 CH954181 EDV49508.1 AGTP01066622 UFQS01000022 UFQT01000022 SSW97581.1 SSX17967.1
PCG81095.1 PCG81096.1 KQ461198 KPJ06266.1 GDQN01001657 JAT89397.1 KQ459606 KPI91669.1 ODYU01005735 SOQ46892.1 AGBW02014334 OWR41847.1 AK401985 BAM18607.1 GDQN01010887 JAT80167.1 KQ971372 EFA10745.2 KQ435763 KOX75408.1 PYGN01000041 PSN56475.1 HQ828078 HQ828080 AET36540.1 AET36541.1 NNAY01000281 OXU29528.1 KZ288254 PBC30718.1 KQ414668 KOC64588.1 KB632328 ERL92529.1 APGK01043278 APGK01043279 KB741014 ENN75407.1 GEDC01001081 JAS36217.1 GDIP01156349 LRGB01002481 JAJ67053.1 KZS07417.1 GDIP01129164 JAL74550.1 GDIP01211405 JAJ11997.1 GDIP01040173 JAM63542.1 LJIJ01000205 ODN00478.1 GDIP01007204 JAM96511.1 LNIX01000020 OXA43912.1 GDIQ01179334 JAK72391.1 GDIQ01224870 JAK26855.1 GDIQ01234869 JAK16856.1 GDIQ01179333 JAK72392.1 GDIQ01022674 JAN72063.1 GDIP01174153 JAJ49249.1 AMQN01013196 KB310036 ELT92677.1 GFWZ01000120 MBW20110.1 KT754480 ALS04314.1 KQ971315 EEZ99309.1 KT754479 KT754481 ALS04315.1 GL447768 EFN85977.1 DQ979802 ABL09307.1 KT754478 ALS04312.1 QCYY01003397 ROT63697.1 GALX01000698 JAB67768.1 KT754735 ALS04569.1 KT755010 ALS04844.1 KT755063 ALS04897.1 BDGG01000001 GAU88111.1 ADTU01006917 GFAH01000229 JAV48160.1 AK416948 BAN20163.1 KT755062 ALS04896.1 DQ979808 ABL09313.1 LBMM01005278 KMQ91658.1 AZHF01000004 OAA76109.1 AK416989 BAN20204.1 EU106629 ABU97480.1 CH477369 EAT42473.1 GALA01000600 JAA94252.1 AY710432 AAU11502.1 GEZM01024990 JAV87652.1 CH954181 EDV49508.1 AGTP01066622 UFQS01000022 UFQT01000022 SSW97581.1 SSX17967.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000053105 UP000245037 UP000005203 UP000215335 UP000002358 UP000192223 UP000242457 UP000053825 UP000030742 UP000019118 UP000076858 UP000094527 UP000198287 UP000014760 UP000008237 UP000283509 UP000186922 UP000005205 UP000036403 UP000076881 UP000008820 UP000008711 UP000005215
UP000007266 UP000053105 UP000245037 UP000005203 UP000215335 UP000002358 UP000192223 UP000242457 UP000053825 UP000030742 UP000019118 UP000076858 UP000094527 UP000198287 UP000014760 UP000008237 UP000283509 UP000186922 UP000005205 UP000036403 UP000076881 UP000008820 UP000008711 UP000005215
PRIDE
Pfam
PF00061 Lipocalin
Interpro
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9JB38
S4PR71
A0A3S2L9C7
A0A2A4K9X7
A0A2A4KAT4
A0A194QRU9
+ More
A0A1E1WR56 A0A194PG40 A0A2H1W239 A0A212EK27 I4DL17 A0A1E1VZL1 D6X2H6 A0A0N1ITP0 A0A2P8ZJ00 A0A087ZPP1 S4S5K9 A0A232FGE3 K7IQ80 A0A1W4WFN5 A0A2A3EG77 A0A0L7R166 U4UEX6 N6U9W4 A0A1B6EE81 A0A0P5DJW0 A0A0P5T9I6 A0A0P4ZWH9 A0A0P6AP56 A0A1D2N5D4 A0A0P6D1A5 A0A226DEB1 A0A0P5L708 A0A0P5HPX8 A0A0P5IR43 A0A0P5KTU4 A0A0P6H9Q8 A0A0N8A7S3 R7TGQ9 A0A2I9LNZ4 A0A0U2KD12 D6WBV4 A0A0U2UEZ0 E2BED2 B0KZJ6 A0A0U2LEV9 A0A3R7LUW6 V5H4N0 A0A0U2TK48 A0A0U2V2N8 A0A0U2T5Y2 A0A1D1UF68 A0A158P1Y8 A0A1W7RAI2 R4WCK5 A0A0U2T8A5 B0KZK2 A0A0J7KMQ5 A0A162N623 R4WCU0 A7XZL4 Q177Y4 T1E383 Q66RP5 A0A1Y1MPR8 B3P4J3 I3MNX9 A0A336JZJ3
A0A1E1WR56 A0A194PG40 A0A2H1W239 A0A212EK27 I4DL17 A0A1E1VZL1 D6X2H6 A0A0N1ITP0 A0A2P8ZJ00 A0A087ZPP1 S4S5K9 A0A232FGE3 K7IQ80 A0A1W4WFN5 A0A2A3EG77 A0A0L7R166 U4UEX6 N6U9W4 A0A1B6EE81 A0A0P5DJW0 A0A0P5T9I6 A0A0P4ZWH9 A0A0P6AP56 A0A1D2N5D4 A0A0P6D1A5 A0A226DEB1 A0A0P5L708 A0A0P5HPX8 A0A0P5IR43 A0A0P5KTU4 A0A0P6H9Q8 A0A0N8A7S3 R7TGQ9 A0A2I9LNZ4 A0A0U2KD12 D6WBV4 A0A0U2UEZ0 E2BED2 B0KZJ6 A0A0U2LEV9 A0A3R7LUW6 V5H4N0 A0A0U2TK48 A0A0U2V2N8 A0A0U2T5Y2 A0A1D1UF68 A0A158P1Y8 A0A1W7RAI2 R4WCK5 A0A0U2T8A5 B0KZK2 A0A0J7KMQ5 A0A162N623 R4WCU0 A7XZL4 Q177Y4 T1E383 Q66RP5 A0A1Y1MPR8 B3P4J3 I3MNX9 A0A336JZJ3
PDB
3PP6
E-value=8.66722e-11,
Score=154
Ontologies
GO
PANTHER
Topology
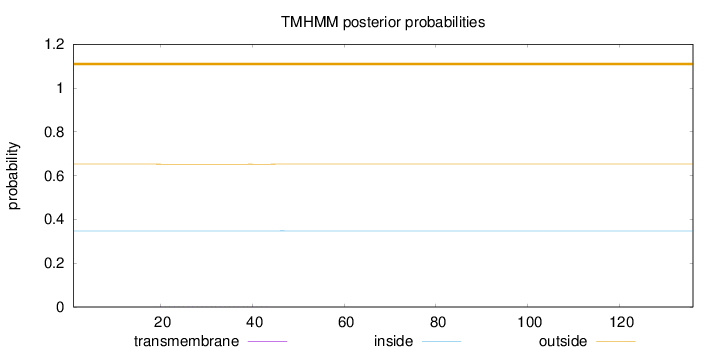
Length:
136
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0165
Exp number, first 60 AAs:
0.0165
Total prob of N-in:
0.34736
outside
1 - 136
Population Genetic Test Statistics
Pi
188.802304
Theta
188.340641
Tajima's D
0.082142
CLR
166.800297
CSRT
0.385430728463577
Interpretation
Possibly Positive selection
