Gene
KWMTBOMO05523
Pre Gene Modal
BGIBMGA006612
Annotation
PREDICTED:_protein_piccolo-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.073
Sequence
CDS
ATGAAATCCTACAACACAAACCTCACGGAATTTTATATAGCGCTGAGACTGATGCAAGCTGTGACAGCCCGTGTAAACAATGTATGCTTGCGGTACTTAGATAATTCCAAGCTTGACACACTCCCTCCCCCACCTCCTATCGCACAAAGAGATCTCAAGACTGTTTCATGTGCCACCAAAAACGGTAGGAGAGTTATGGAAGATAGACACGTTGAATTCGGAAATTTGGAGGCACTGTTTGGAAAAGAAACGACAGAGCCTACAAGTTTCTATGCAGTGTATGATGGGCATGCAGGCTCGGCCGCGGCGACTTATTGCGCGGCGCATTTACACCAGTACCTCATTGAGAGCCCACATTTCACTACTGACCTACAACGAGCGATGCGAGACGCCTTCTTAAGAACCGATGCAGAATTTATCAGGAAAAGTAATCAAGAGCGAGCGAACGGTGGCAGCACGGCGGTGGCGGTGTGCCTGCGCGGGTCGCAGCTGCTGGCGGCCTGGGCGGGCGACTCGCTGGCGCTGCTCGCCAAGCGCATGCGGCTCATGCAGCTCGTGCGCCCGCACAAACCGGGACGACAGGACGAACGAGATCGGATAGAGACAAGCGGAGGGACAGTAATCTACTGGGGCACGTGGCGAGTCAACGGTCAATTGGCAGTTTCCAGGGCCATTGGTGACGCACAATACAAGCCGTACGTGACGGCGCGGCCCGAGATCGCGACAGTGGACCTCGACGGCGACGAAGACTTCGTTGTGATTGCTTGCGACGGACTCTGGGACTTTGTTACCGAAGACGACGTCGCCCTCTCCGTTTATAGGCAGCTCGCCGCCGATCCGGGTAAGTCTACGAATACACGCTTAAATACATACTAG
Protein
MKSYNTNLTEFYIALRLMQAVTARVNNVCLRYLDNSKLDTLPPPPPIAQRDLKTVSCATKNGRRVMEDRHVEFGNLEALFGKETTEPTSFYAVYDGHAGSAAATYCAAHLHQYLIESPHFTTDLQRAMRDAFLRTDAEFIRKSNQERANGGSTAVAVCLRGSQLLAAWAGDSLALLAKRMRLMQLVRPHKPGRQDERDRIETSGGTVIYWGTWRVNGQLAVSRAIGDAQYKPYVTARPEIATVDLDGDEDFVVIACDGLWDFVTEDDVALSVYRQLAADPGKSTNTRLNTY
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the PP2C family.
Uniprot
H9JAR7
A0A2H1W1K0
A0A194QL56
A0A194QJJ9
A0A212EK76
A0A2J7RBD1
+ More
A0A139WB92 A0A139WB93 A0A1W4WFM3 A0A1W4W3T7 A0A1W4WEC7 A0A1W4WFC1 A0A1W4W3S7 A0A067QXB8 E2AHG4 F4W662 U4U0S6 N6U9W8 A0A151WT37 A0A195AW93 A0A151IKJ0 E2BEC9 A0A3L8DQT4 A0A195DMM5 A0A1B6GB92 A0A0C9R2P2 A0A1Y1NAQ2 A0A158P2K5 A0A1B6IUI9 A0A0N8EAC8 A0A1B6LH02 A0A0P5YB07 A0A0P5IG42 A0A0P5GJC3 A0A0P5YB74 A0A0P6ELH1 A0A232FFI8 A0A0P5SKK9 A0A0P6FWB2 A0A0N8EFM7 A0A087ZQ25 A0A0P5XSZ9 A0A0P5WS52 A0A0N8D4H1 A0A0P4WPP8 A0A0P5UMG5 A0A0P5BBB9 A0A0P4XNC0 A0A0P5E1K3 A0A0P5UMY3 A0A0P5AGV6 A0A0P5YA24 A0A0P5WHS2 A0A0P5X959 A0A0P5PPP2 A0A0P6B8F5 A0A0P5I0S2 A0A0P6EA63 A0A1B6DD99 A0A0P6EVR0 A0A0P6GQ86 A0A0P6EFW4 A0A0P6FJF7 A0A0P6FYV2 A0A076FEQ3 A0A0P5GXV1 A0A0N8C3W8 A0A0N8C9T5 A0A0P5PQI0 A0A0P5I5Z9 A0A0P5HYB0 A0A0P4YQJ9 A0A0P5HP47 A0A0P5HK57 A0A0P5IBX9 A0A0N8B727 A0A0P5HMZ4 A0A0P5HVQ1 A0A0P5KCD6 A0A0P5KQE1 A0A0P5HN99 A0A0P5ES46 A0A0P5KKC2 A0A0P5KLU6 A0A0P5I9A5 A0A0P5KCD9 A0A0P5I2Y8 A0A195FJB2 A0A0P5KR49 A0A0N8BUD8 A0A0P5IF77 A0A0P5HR02 A0A0P5HSC3 A0A0P5KRF6 A0A0P5K5T2 A0A0P5JEM9 A0A0P5J2F3 A0A0P5HWF5
A0A139WB92 A0A139WB93 A0A1W4WFM3 A0A1W4W3T7 A0A1W4WEC7 A0A1W4WFC1 A0A1W4W3S7 A0A067QXB8 E2AHG4 F4W662 U4U0S6 N6U9W8 A0A151WT37 A0A195AW93 A0A151IKJ0 E2BEC9 A0A3L8DQT4 A0A195DMM5 A0A1B6GB92 A0A0C9R2P2 A0A1Y1NAQ2 A0A158P2K5 A0A1B6IUI9 A0A0N8EAC8 A0A1B6LH02 A0A0P5YB07 A0A0P5IG42 A0A0P5GJC3 A0A0P5YB74 A0A0P6ELH1 A0A232FFI8 A0A0P5SKK9 A0A0P6FWB2 A0A0N8EFM7 A0A087ZQ25 A0A0P5XSZ9 A0A0P5WS52 A0A0N8D4H1 A0A0P4WPP8 A0A0P5UMG5 A0A0P5BBB9 A0A0P4XNC0 A0A0P5E1K3 A0A0P5UMY3 A0A0P5AGV6 A0A0P5YA24 A0A0P5WHS2 A0A0P5X959 A0A0P5PPP2 A0A0P6B8F5 A0A0P5I0S2 A0A0P6EA63 A0A1B6DD99 A0A0P6EVR0 A0A0P6GQ86 A0A0P6EFW4 A0A0P6FJF7 A0A0P6FYV2 A0A076FEQ3 A0A0P5GXV1 A0A0N8C3W8 A0A0N8C9T5 A0A0P5PQI0 A0A0P5I5Z9 A0A0P5HYB0 A0A0P4YQJ9 A0A0P5HP47 A0A0P5HK57 A0A0P5IBX9 A0A0N8B727 A0A0P5HMZ4 A0A0P5HVQ1 A0A0P5KCD6 A0A0P5KQE1 A0A0P5HN99 A0A0P5ES46 A0A0P5KKC2 A0A0P5KLU6 A0A0P5I9A5 A0A0P5KCD9 A0A0P5I2Y8 A0A195FJB2 A0A0P5KR49 A0A0N8BUD8 A0A0P5IF77 A0A0P5HR02 A0A0P5HSC3 A0A0P5KRF6 A0A0P5K5T2 A0A0P5JEM9 A0A0P5J2F3 A0A0P5HWF5
Pubmed
EMBL
BABH01008535
ODYU01005735
SOQ46893.1
KQ461198
KPJ06267.1
KQ458793
+ More
KPJ05090.1 AGBW02014334 OWR41848.1 NEVH01005906 PNF38145.1 KQ971372 KYB25190.1 KYB25189.1 KK853247 KDR09383.1 GL439539 EFN67119.1 GL887707 EGI70197.1 KB631252 KB632328 ERL84191.1 ERL92524.1 APGK01043279 KB741014 ENN75412.1 KQ982763 KYQ51014.1 KQ976731 KYM76305.1 KQ977199 KYN04988.1 GL447768 EFN85974.1 QOIP01000005 RLU22632.1 KQ980724 KYN14128.1 GECZ01010061 JAS59708.1 GBYB01010429 JAG80196.1 GEZM01011966 JAV93326.1 ADTU01006915 ADTU01006916 ADTU01006917 GECU01017118 JAS90588.1 GDIQ01046244 JAN48493.1 GEBQ01017002 JAT22975.1 GDIP01060281 JAM43434.1 GDIQ01221775 JAK29950.1 GDIQ01247070 JAK04655.1 GDIP01060282 JAM43433.1 GDIQ01060989 JAN33748.1 NNAY01000281 OXU29524.1 GDIQ01094472 JAL57254.1 GDIQ01041899 JAN52838.1 GDIQ01031452 JAN63285.1 GDIP01069678 JAM34037.1 GDIP01083275 JAM20440.1 GDIP01069679 JAM34036.1 GDIP01253403 JAI69998.1 GDIP01110868 JAL92846.1 GDIP01186860 LRGB01000568 JAJ36542.1 KZS17823.1 GDIP01239259 JAI84142.1 GDIP01148372 JAJ75030.1 GDIP01110869 JAL92845.1 GDIP01199015 JAJ24387.1 GDIP01062236 JAM41479.1 GDIP01086325 JAM17390.1 GDIP01075524 JAM28191.1 GDIQ01125212 JAL26514.1 GDIP01020157 JAM83558.1 GDIQ01220378 JAK31347.1 GDIQ01077525 JAN17212.1 GEDC01013676 JAS23622.1 GDIQ01064584 JAN30153.1 GDIQ01030360 JAN64377.1 GDIQ01077526 JAN17211.1 GDIQ01058838 JAN35899.1 GDIQ01058837 JAN35900.1 KF516594 AII16498.1 GDIQ01241021 JAK10704.1 GDIQ01117315 JAL34411.1 GDIQ01100779 JAL50947.1 GDIQ01126918 JAL24808.1 GDIQ01241022 JAK10703.1 GDIQ01223228 JAK28497.1 GDIP01234556 JAI88845.1 GDIQ01226830 JAK24895.1 GDIQ01226829 JAK24896.1 GDIQ01216983 JAK34742.1 GDIQ01206443 JAK45282.1 GDIQ01225677 JAK26048.1 GDIQ01224480 JAK27245.1 GDIQ01186103 JAK65622.1 GDIQ01206444 JAK45281.1 GDIQ01225676 GDIQ01225675 GDIQ01225674 JAK26051.1 GDIQ01268296 JAJ83428.1 GDIQ01184342 JAK67383.1 GDIQ01207797 JAK43928.1 GDIQ01216982 JAK34743.1 GDIQ01187697 GDIQ01187695 GDIQ01187694 JAK64028.1 GDIQ01224481 GDIQ01224479 GDIQ01224478 JAK27247.1 KQ981523 KYN40456.1 GDIQ01187696 JAK64029.1 GDIQ01143955 JAL07771.1 GDIQ01218078 JAK33647.1 GDIQ01224477 JAK27248.1 GDIQ01225678 JAK26047.1 GDIQ01187693 JAK64032.1 GDIQ01190337 JAK61388.1 GDIQ01206442 JAK45283.1 GDIQ01206441 JAK45284.1 GDIQ01226828 JAK24897.1
KPJ05090.1 AGBW02014334 OWR41848.1 NEVH01005906 PNF38145.1 KQ971372 KYB25190.1 KYB25189.1 KK853247 KDR09383.1 GL439539 EFN67119.1 GL887707 EGI70197.1 KB631252 KB632328 ERL84191.1 ERL92524.1 APGK01043279 KB741014 ENN75412.1 KQ982763 KYQ51014.1 KQ976731 KYM76305.1 KQ977199 KYN04988.1 GL447768 EFN85974.1 QOIP01000005 RLU22632.1 KQ980724 KYN14128.1 GECZ01010061 JAS59708.1 GBYB01010429 JAG80196.1 GEZM01011966 JAV93326.1 ADTU01006915 ADTU01006916 ADTU01006917 GECU01017118 JAS90588.1 GDIQ01046244 JAN48493.1 GEBQ01017002 JAT22975.1 GDIP01060281 JAM43434.1 GDIQ01221775 JAK29950.1 GDIQ01247070 JAK04655.1 GDIP01060282 JAM43433.1 GDIQ01060989 JAN33748.1 NNAY01000281 OXU29524.1 GDIQ01094472 JAL57254.1 GDIQ01041899 JAN52838.1 GDIQ01031452 JAN63285.1 GDIP01069678 JAM34037.1 GDIP01083275 JAM20440.1 GDIP01069679 JAM34036.1 GDIP01253403 JAI69998.1 GDIP01110868 JAL92846.1 GDIP01186860 LRGB01000568 JAJ36542.1 KZS17823.1 GDIP01239259 JAI84142.1 GDIP01148372 JAJ75030.1 GDIP01110869 JAL92845.1 GDIP01199015 JAJ24387.1 GDIP01062236 JAM41479.1 GDIP01086325 JAM17390.1 GDIP01075524 JAM28191.1 GDIQ01125212 JAL26514.1 GDIP01020157 JAM83558.1 GDIQ01220378 JAK31347.1 GDIQ01077525 JAN17212.1 GEDC01013676 JAS23622.1 GDIQ01064584 JAN30153.1 GDIQ01030360 JAN64377.1 GDIQ01077526 JAN17211.1 GDIQ01058838 JAN35899.1 GDIQ01058837 JAN35900.1 KF516594 AII16498.1 GDIQ01241021 JAK10704.1 GDIQ01117315 JAL34411.1 GDIQ01100779 JAL50947.1 GDIQ01126918 JAL24808.1 GDIQ01241022 JAK10703.1 GDIQ01223228 JAK28497.1 GDIP01234556 JAI88845.1 GDIQ01226830 JAK24895.1 GDIQ01226829 JAK24896.1 GDIQ01216983 JAK34742.1 GDIQ01206443 JAK45282.1 GDIQ01225677 JAK26048.1 GDIQ01224480 JAK27245.1 GDIQ01186103 JAK65622.1 GDIQ01206444 JAK45281.1 GDIQ01225676 GDIQ01225675 GDIQ01225674 JAK26051.1 GDIQ01268296 JAJ83428.1 GDIQ01184342 JAK67383.1 GDIQ01207797 JAK43928.1 GDIQ01216982 JAK34743.1 GDIQ01187697 GDIQ01187695 GDIQ01187694 JAK64028.1 GDIQ01224481 GDIQ01224479 GDIQ01224478 JAK27247.1 KQ981523 KYN40456.1 GDIQ01187696 JAK64029.1 GDIQ01143955 JAL07771.1 GDIQ01218078 JAK33647.1 GDIQ01224477 JAK27248.1 GDIQ01225678 JAK26047.1 GDIQ01187693 JAK64032.1 GDIQ01190337 JAK61388.1 GDIQ01206442 JAK45283.1 GDIQ01206441 JAK45284.1 GDIQ01226828 JAK24897.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF81606
SSF81606
Gene 3D
CDD
ProteinModelPortal
H9JAR7
A0A2H1W1K0
A0A194QL56
A0A194QJJ9
A0A212EK76
A0A2J7RBD1
+ More
A0A139WB92 A0A139WB93 A0A1W4WFM3 A0A1W4W3T7 A0A1W4WEC7 A0A1W4WFC1 A0A1W4W3S7 A0A067QXB8 E2AHG4 F4W662 U4U0S6 N6U9W8 A0A151WT37 A0A195AW93 A0A151IKJ0 E2BEC9 A0A3L8DQT4 A0A195DMM5 A0A1B6GB92 A0A0C9R2P2 A0A1Y1NAQ2 A0A158P2K5 A0A1B6IUI9 A0A0N8EAC8 A0A1B6LH02 A0A0P5YB07 A0A0P5IG42 A0A0P5GJC3 A0A0P5YB74 A0A0P6ELH1 A0A232FFI8 A0A0P5SKK9 A0A0P6FWB2 A0A0N8EFM7 A0A087ZQ25 A0A0P5XSZ9 A0A0P5WS52 A0A0N8D4H1 A0A0P4WPP8 A0A0P5UMG5 A0A0P5BBB9 A0A0P4XNC0 A0A0P5E1K3 A0A0P5UMY3 A0A0P5AGV6 A0A0P5YA24 A0A0P5WHS2 A0A0P5X959 A0A0P5PPP2 A0A0P6B8F5 A0A0P5I0S2 A0A0P6EA63 A0A1B6DD99 A0A0P6EVR0 A0A0P6GQ86 A0A0P6EFW4 A0A0P6FJF7 A0A0P6FYV2 A0A076FEQ3 A0A0P5GXV1 A0A0N8C3W8 A0A0N8C9T5 A0A0P5PQI0 A0A0P5I5Z9 A0A0P5HYB0 A0A0P4YQJ9 A0A0P5HP47 A0A0P5HK57 A0A0P5IBX9 A0A0N8B727 A0A0P5HMZ4 A0A0P5HVQ1 A0A0P5KCD6 A0A0P5KQE1 A0A0P5HN99 A0A0P5ES46 A0A0P5KKC2 A0A0P5KLU6 A0A0P5I9A5 A0A0P5KCD9 A0A0P5I2Y8 A0A195FJB2 A0A0P5KR49 A0A0N8BUD8 A0A0P5IF77 A0A0P5HR02 A0A0P5HSC3 A0A0P5KRF6 A0A0P5K5T2 A0A0P5JEM9 A0A0P5J2F3 A0A0P5HWF5
A0A139WB92 A0A139WB93 A0A1W4WFM3 A0A1W4W3T7 A0A1W4WEC7 A0A1W4WFC1 A0A1W4W3S7 A0A067QXB8 E2AHG4 F4W662 U4U0S6 N6U9W8 A0A151WT37 A0A195AW93 A0A151IKJ0 E2BEC9 A0A3L8DQT4 A0A195DMM5 A0A1B6GB92 A0A0C9R2P2 A0A1Y1NAQ2 A0A158P2K5 A0A1B6IUI9 A0A0N8EAC8 A0A1B6LH02 A0A0P5YB07 A0A0P5IG42 A0A0P5GJC3 A0A0P5YB74 A0A0P6ELH1 A0A232FFI8 A0A0P5SKK9 A0A0P6FWB2 A0A0N8EFM7 A0A087ZQ25 A0A0P5XSZ9 A0A0P5WS52 A0A0N8D4H1 A0A0P4WPP8 A0A0P5UMG5 A0A0P5BBB9 A0A0P4XNC0 A0A0P5E1K3 A0A0P5UMY3 A0A0P5AGV6 A0A0P5YA24 A0A0P5WHS2 A0A0P5X959 A0A0P5PPP2 A0A0P6B8F5 A0A0P5I0S2 A0A0P6EA63 A0A1B6DD99 A0A0P6EVR0 A0A0P6GQ86 A0A0P6EFW4 A0A0P6FJF7 A0A0P6FYV2 A0A076FEQ3 A0A0P5GXV1 A0A0N8C3W8 A0A0N8C9T5 A0A0P5PQI0 A0A0P5I5Z9 A0A0P5HYB0 A0A0P4YQJ9 A0A0P5HP47 A0A0P5HK57 A0A0P5IBX9 A0A0N8B727 A0A0P5HMZ4 A0A0P5HVQ1 A0A0P5KCD6 A0A0P5KQE1 A0A0P5HN99 A0A0P5ES46 A0A0P5KKC2 A0A0P5KLU6 A0A0P5I9A5 A0A0P5KCD9 A0A0P5I2Y8 A0A195FJB2 A0A0P5KR49 A0A0N8BUD8 A0A0P5IF77 A0A0P5HR02 A0A0P5HSC3 A0A0P5KRF6 A0A0P5K5T2 A0A0P5JEM9 A0A0P5J2F3 A0A0P5HWF5
PDB
4OIC
E-value=1.01048e-30,
Score=331
Ontologies
GO
PANTHER
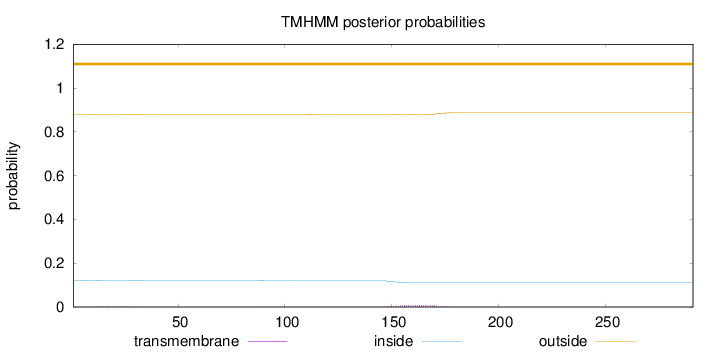
Topology
Length:
291
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25036
Exp number, first 60 AAs:
0.03563
Total prob of N-in:
0.12148
outside
1 - 291
Population Genetic Test Statistics
Pi
211.995348
Theta
175.335802
Tajima's D
0.629281
CLR
0.173373
CSRT
0.552872356382181
Interpretation
Possibly Positive selection
