Gene
KWMTBOMO05514
Pre Gene Modal
BGIBMGA006726
Annotation
death_related_ced-3/Nedd2-like_protein_[Bombyx_mori]
Full name
Caspase-8
Alternative Name
Death-related ced-3/NEDD2-like protein
Location in the cell
Nuclear Reliability : 3.245
Sequence
CDS
ATGTTTCGACCTGACGCTTTAGATGAACGAGCTGCATTAGACCGACAGATTATTCACAGCAATCTTATAAATGTCGATGTCATTTCTCAAATCGAAAGAGAATTGCAAGATGAGCCGTATGACATGGTATCTCTAGTGTTCCTGTTGTATGAAGTACCGGATACTGCTTTGCAGAAGTTAGTTACACATCAAAAGATAGTGACAGAAATGCTTGGAACTAATTTGAATTTACTTCATGACTGGTATCAGCATTCAAAATCGAAACCCACTTGGAAACATGAGTTCCTGGAGGCACTTCTTATTTGTCAATTGTTTAACATAGTTCGAAGAATAGGATTTGATGTCCAAACGCTTCGAAAGCATTACCAGACAGATTACCCAGGATTAAGTATGTATGTTGATCCTTTAAGAAAGATTTTATATAAAATATGTGAAGAAATAGACACTCCAAATTTAATAAAACTTCAAAAATCCTTATTGACATATGATATAGATGTAAGCGGGCACAATATATGTGAAATTATATTACTTGAACTTATGTCAAGAAGATTCATAGGGATTAAGTATTACAGACATGATGAAAAATACCTTACTGAAATAAAAATTGATAAGTTTCTGAGAATAATTGAAAACTTTGATGGATTGAGAAAGTTATCATTAGATTTAAAATTTCTACAGAATAAATTTGCCAATGAAACAAATCCAAATAATTCCCTCAAGTGTAAGATGGATACTACATCACAACAAAATGAACAACCTGTATTAGTTGAACATACAAATGAAGGAGGGATTTTTGATATTAACAATCTTTTAGACATACATGAAGATTTAGAAAAATTGCAAATTAATGATGGTTTTCATTTTGAAGCCGATAAGAATAGACACATGGAAGAAATTTATGAAATAAAAAATAAAAATAGATTAGGATTGTGTTTAATAATAAATCAAGAGAACTTCTATCCATCAAGGCAAAGTATAGAACTTAACAATCAAATGGATCCATTACAAACAAGGACTGGATCAACCAAAGACAGAATTGCATTAGAAAATACAATGTCATTATTTAATTTCGTAGTTAATAGTCGTTCTGACTTGGGACACAAAGAGATGATTAATTTCATAAAAGATAATATCAAAAACAATATTCATGAAGATGATAGCATGTTCATGTTATGTATTTTGTCACATGGTGTGCGAGGGCACATTTTTGCAGCAGATTCAGTGAAAATTAAAGTTGAAGATATACAAAAGCTATTGGACTCTGATGAAGCAAAGAAACTGCATTGCATACCGAAAGTTCTTATAATCCAAGCTTGTCAAGTTATTGAAAACCAAAATCCATATCTAACAACTGACTCGCCCTTGGGTAATATATTCATAAAAAAATCAGATTTTCTAGTATACTGGGCAACAGCACCTGAATATGAAGCTATAAGACACATTACTAAAGGAACTGTGTTCATACAAAATCTTTGCAAAGCAATTAGACTTTATGGAAATAAAAAACATTTATCAGATATTTTTACTATTGTCAATAATACAGTCACAAACTACTGCATGAAACTGAATCATCCTCAGGTTCCCATTTATGAAAATAGCTTAAGAAGGGCACTTTACTTATTTAAGCCATGA
Protein
MFRPDALDERAALDRQIIHSNLINVDVISQIERELQDEPYDMVSLVFLLYEVPDTALQKLVTHQKIVTEMLGTNLNLLHDWYQHSKSKPTWKHEFLEALLICQLFNIVRRIGFDVQTLRKHYQTDYPGLSMYVDPLRKILYKICEEIDTPNLIKLQKSLLTYDIDVSGHNICEIILLELMSRRFIGIKYYRHDEKYLTEIKIDKFLRIIENFDGLRKLSLDLKFLQNKFANETNPNNSLKCKMDTTSQQNEQPVLVEHTNEGGIFDINNLLDIHEDLEKLQINDGFHFEADKNRHMEEIYEIKNKNRLGLCLIINQENFYPSRQSIELNNQMDPLQTRTGSTKDRIALENTMSLFNFVVNSRSDLGHKEMINFIKDNIKNNIHEDDSMFMLCILSHGVRGHIFAADSVKIKVEDIQKLLDSDEAKKLHCIPKVLIIQACQVIENQNPYLTTDSPLGNIFIKKSDFLVYWATAPEYEAIRHITKGTVFIQNLCKAIRLYGNKKHLSDIFTIVNNTVTNYCMKLNHPQVPIYENSLRRALYLFKP
Summary
Description
Effector of the programmed cell death (PCD) activators rpr, grim and W. May play an apoptotic role in the germline as well as soma. Role in immune response, required to resist Gram-negative bacterial infections by regulating DptA. Fadd interacts with Dredd, Fadd promotes cleavage of Dredd and is necessary and sufficient for enhancing Dredd-induced apoptosis.
Effector of the programmed cell death (PCD) activators rpr, grim and hid (PubMed:9740659). May play an apoptotic role in the germline as well as soma. Fadd interacts with Dredd to promote cleavage of Dredd and is necessary and sufficient for enhancing Dredd-induced apoptosis (PubMed:10934188). Plays a role in the innate immune response. Required for resistance to Gram-negative bacterial infection (PubMed:11269502). Diap2-mediated ubiquitination of Dredd is critical for processing of imd and rel and the subsequent expression of antimicrobial genes such as DptA (PubMed:22549468).
Effector of the programmed cell death (PCD) activators rpr, grim and hid (PubMed:9740659). May play an apoptotic role in the germline as well as soma. Fadd interacts with Dredd to promote cleavage of Dredd and is necessary and sufficient for enhancing Dredd-induced apoptosis (PubMed:10934188). Plays a role in the innate immune response. Required for resistance to Gram-negative bacterial infection (PubMed:11269502). Diap2-mediated ubiquitination of Dredd is critical for processing of imd and rel and the subsequent expression of antimicrobial genes such as DptA (PubMed:22549468).
Catalytic Activity
Strict requirement for Asp at position P1 and has a preferred cleavage sequence of (Leu/Asp/Val)-Glu-Thr-Asp-|-(Gly/Ser/Ala).
Subunit
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 15 kDa (caspase-8 subunit p15) and a 10 kDa (caspase-8 subunit p10) subunit. Interacts with the N-terminus of Fadd (By similarity).
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 15 kDa (caspase-8 subunit p15) and a 10 kDa (caspase-8 subunit p10) subunit (PubMed:9740659). Interacts (via N-terminus) with Diap2; likely to bind Diap2 simultaneously with Fadd to form a trimeric complex (PubMed:10934188, PubMed:22549468).
Heterotetramer that consists of two anti-parallel arranged heterodimers, each one formed by a 15 kDa (caspase-8 subunit p15) and a 10 kDa (caspase-8 subunit p10) subunit (PubMed:9740659). Interacts (via N-terminus) with Diap2; likely to bind Diap2 simultaneously with Fadd to form a trimeric complex (PubMed:10934188, PubMed:22549468).
Similarity
Belongs to the peptidase C14A family.
Keywords
Apoptosis
Complete proteome
Cytoplasm
Hydrolase
Immunity
Innate immunity
Protease
Reference proteome
Thiol protease
Zymogen
Alternative splicing
Ubl conjugation
Feature
chain Caspase-8 subunit p15
splice variant In isoform alpha.
splice variant In isoform alpha.
Uniprot
H9JB31
A9ZMY8
A0A2A4KAV9
G0XQH8
G0XQH7
I7CAH9
+ More
A0A142J2X8 A0A0L7LPH4 R4I3K4 G0XQH9 F6K5S2 A0A194QL32 A0A1E1WQP3 A0A212FDA5 A0A194QJI1 G0XQH6 A0A346RAD7 D6WJH5 A0A3Q8C047 V5GYQ4 A0A3F2Z153 A0A1W4W5I4 A0A1W4WH04 A0A1W4W5J4 A0A182N2I9 A0A182HGZ6 A0A182L4D4 A0A182QBD6 A0A182VMT7 Q16H55 A0A1S4H7C5 W8AV04 A0A3F2YVF2 A3QVU6 A0A067QV28 A0A182SF59 W5JJK4 A0A084WBG7 A0A3F2YXQ1 A0A1B0D025 A0A182FTB8 A0A3F2YU15 A0A3F2YWL0 Q29IM7 A0A0R3NZ05 A0A0R3NYQ6 A0A1B0GLN8 B4M7Y9 A0A1I8M1X4 A0A0D3QBR4 B4N271 A0A0D3QC48 A0A0D3QBU5 A0A0D3QBU2 A0A0D3QBS0 A0A0D3QBQ6 A0A0D3QBR1 A0A1A9WRC9 A0A0D3QBR0 A0A0A1XM78 A0A0D3QC59 A0A0D3QBR6 A0A1Y1MQM3 A0A182H1W5 B4PX19 A0A1I8PI09 A0A0D3QCD4 A0A0M4EZM9 N6TGN6 B3P987 A0A0K8TMN0 B4JNA9 Q8IRY7 X2JHT0 A0A0L0CNR9 A0A336MAB9 A0A3Q9HGP0 A0A1B6JA75 B4H2Q9 A0A1B6HSW3 A0A0K1DH04 A0A1B0ALP5
A0A142J2X8 A0A0L7LPH4 R4I3K4 G0XQH9 F6K5S2 A0A194QL32 A0A1E1WQP3 A0A212FDA5 A0A194QJI1 G0XQH6 A0A346RAD7 D6WJH5 A0A3Q8C047 V5GYQ4 A0A3F2Z153 A0A1W4W5I4 A0A1W4WH04 A0A1W4W5J4 A0A182N2I9 A0A182HGZ6 A0A182L4D4 A0A182QBD6 A0A182VMT7 Q16H55 A0A1S4H7C5 W8AV04 A0A3F2YVF2 A3QVU6 A0A067QV28 A0A182SF59 W5JJK4 A0A084WBG7 A0A3F2YXQ1 A0A1B0D025 A0A182FTB8 A0A3F2YU15 A0A3F2YWL0 Q29IM7 A0A0R3NZ05 A0A0R3NYQ6 A0A1B0GLN8 B4M7Y9 A0A1I8M1X4 A0A0D3QBR4 B4N271 A0A0D3QC48 A0A0D3QBU5 A0A0D3QBU2 A0A0D3QBS0 A0A0D3QBQ6 A0A0D3QBR1 A0A1A9WRC9 A0A0D3QBR0 A0A0A1XM78 A0A0D3QC59 A0A0D3QBR6 A0A1Y1MQM3 A0A182H1W5 B4PX19 A0A1I8PI09 A0A0D3QCD4 A0A0M4EZM9 N6TGN6 B3P987 A0A0K8TMN0 B4JNA9 Q8IRY7 X2JHT0 A0A0L0CNR9 A0A336MAB9 A0A3Q9HGP0 A0A1B6JA75 B4H2Q9 A0A1B6HSW3 A0A0K1DH04 A0A1B0ALP5
EC Number
3.4.22.61
Pubmed
19121390
21740565
26977926
26227816
26354079
22118469
+ More
29910977 18362917 19820115 20966253 17510324 12364791 24495485 17517333 24845553 20920257 23761445 24438588 15632085 17994087 25315136 25552603 25830018 28004739 26483478 17550304 23537049 26369729 9740659 10731132 12537572 10731137 12537569 9380701 11269502 10934188 22549468 12537568 12537573 12537574 16110336 17569856 17569867 26108605 29989657 26073458
29910977 18362917 19820115 20966253 17510324 12364791 24495485 17517333 24845553 20920257 23761445 24438588 15632085 17994087 25315136 25552603 25830018 28004739 26483478 17550304 23537049 26369729 9740659 10731132 12537572 10731137 12537569 9380701 11269502 10934188 22549468 12537568 12537573 12537574 16110336 17569856 17569867 26108605 29989657 26073458
EMBL
BABH01008520
AB292816
BAF98475.1
NWSH01000001
PCG81126.1
HQ328982
+ More
AEK20839.1 HQ328981 AEK20838.1 JX040310 AFO64608.1 KU668855 AMR71144.1 JTDY01000408 KOB77332.1 JQ768052 AFJ04536.1 HQ328983 AEK20840.1 HM234679 AEF30497.1 KQ461198 KPJ06283.1 GDQN01001913 JAT89141.1 AGBW02009100 OWR51688.1 KQ458793 KPJ05070.1 HQ328980 AEK20837.1 MH200927 AXS59137.1 KQ971342 EFA04648.1 KY056148 ATX63070.1 GALX01002893 JAB65573.1 APCN01002368 AXCN02001076 CH478203 EAT33580.1 AAAB01008986 GAMC01016478 JAB90077.1 AXCM01008478 DQ860099 ABI74776.1 KK852914 KDR13883.1 ADMH02001049 ETN64316.1 ATLV01022353 KE525331 KFB47561.1 AJVK01021192 CH379063 EAL32626.1 KRT06185.1 KRT06187.1 AJWK01005641 CH940653 EDW62265.1 KP274213 AJC98276.1 CH963925 EDW78460.1 KP274202 AJC98265.1 KP274219 AJC98282.1 KP274214 AJC98277.1 KP274220 AJC98283.1 KP274203 AJC98266.1 KP274205 AJC98268.1 KP274208 AJC98271.1 GBXI01002230 JAD12062.1 KP274212 AJC98275.1 KP274218 AJC98281.1 GEZM01024088 GEZM01024087 GEZM01024086 JAV87989.1 JXUM01104218 KQ564868 KXJ71641.1 CM000162 EDX00805.2 KP274221 AJC98284.1 CP012528 ALC49680.1 APGK01028352 KB740686 KB631930 ENN79559.1 ERL87316.1 CH954183 EDV45383.2 GDAI01002428 JAI15175.1 CH916371 EDV92202.1 AF007016 AF083894 AF083895 AE014298 AL031581 AY060275 AF031652 AHN59222.1 JRES01000127 KNC33965.1 UFQS01000611 UFQT01000611 SSX05362.1 SSX25723.1 MG279147 AZR38348.1 GECU01011565 JAS96141.1 CH479204 EDW30626.1 GECU01029950 JAS77756.1 KP204031 AKT27901.1 JXJN01000032
AEK20839.1 HQ328981 AEK20838.1 JX040310 AFO64608.1 KU668855 AMR71144.1 JTDY01000408 KOB77332.1 JQ768052 AFJ04536.1 HQ328983 AEK20840.1 HM234679 AEF30497.1 KQ461198 KPJ06283.1 GDQN01001913 JAT89141.1 AGBW02009100 OWR51688.1 KQ458793 KPJ05070.1 HQ328980 AEK20837.1 MH200927 AXS59137.1 KQ971342 EFA04648.1 KY056148 ATX63070.1 GALX01002893 JAB65573.1 APCN01002368 AXCN02001076 CH478203 EAT33580.1 AAAB01008986 GAMC01016478 JAB90077.1 AXCM01008478 DQ860099 ABI74776.1 KK852914 KDR13883.1 ADMH02001049 ETN64316.1 ATLV01022353 KE525331 KFB47561.1 AJVK01021192 CH379063 EAL32626.1 KRT06185.1 KRT06187.1 AJWK01005641 CH940653 EDW62265.1 KP274213 AJC98276.1 CH963925 EDW78460.1 KP274202 AJC98265.1 KP274219 AJC98282.1 KP274214 AJC98277.1 KP274220 AJC98283.1 KP274203 AJC98266.1 KP274205 AJC98268.1 KP274208 AJC98271.1 GBXI01002230 JAD12062.1 KP274212 AJC98275.1 KP274218 AJC98281.1 GEZM01024088 GEZM01024087 GEZM01024086 JAV87989.1 JXUM01104218 KQ564868 KXJ71641.1 CM000162 EDX00805.2 KP274221 AJC98284.1 CP012528 ALC49680.1 APGK01028352 KB740686 KB631930 ENN79559.1 ERL87316.1 CH954183 EDV45383.2 GDAI01002428 JAI15175.1 CH916371 EDV92202.1 AF007016 AF083894 AF083895 AE014298 AL031581 AY060275 AF031652 AHN59222.1 JRES01000127 KNC33965.1 UFQS01000611 UFQT01000611 SSX05362.1 SSX25723.1 MG279147 AZR38348.1 GECU01011565 JAS96141.1 CH479204 EDW30626.1 GECU01029950 JAS77756.1 KP204031 AKT27901.1 JXJN01000032
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000007151
UP000053268
+ More
UP000007266 UP000075920 UP000192223 UP000075884 UP000075840 UP000075882 UP000075886 UP000075903 UP000008820 UP000075883 UP000027135 UP000075901 UP000000673 UP000030765 UP000075900 UP000092462 UP000069272 UP000075881 UP000075885 UP000001819 UP000092461 UP000008792 UP000095301 UP000007798 UP000091820 UP000069940 UP000249989 UP000002282 UP000095300 UP000092553 UP000019118 UP000030742 UP000008711 UP000001070 UP000000803 UP000037069 UP000008744 UP000092460
UP000007266 UP000075920 UP000192223 UP000075884 UP000075840 UP000075882 UP000075886 UP000075903 UP000008820 UP000075883 UP000027135 UP000075901 UP000000673 UP000030765 UP000075900 UP000092462 UP000069272 UP000075881 UP000075885 UP000001819 UP000092461 UP000008792 UP000095301 UP000007798 UP000091820 UP000069940 UP000249989 UP000002282 UP000095300 UP000092553 UP000019118 UP000030742 UP000008711 UP000001070 UP000000803 UP000037069 UP000008744 UP000092460
Interpro
IPR029030
Caspase-like_dom_sf
+ More
IPR001309 Pept_C14_p20
IPR015917 Pept_C14A
IPR002138 Pept_C14_p10
IPR002398 Pept_C14
IPR018484 Carb_kinase_FGGY_N
IPR018485 Carb_kinase_FGGY_C
IPR006003 FGGY_RbtK-like
IPR000352 Pep_chain_release_fac_I_II
IPR033139 Caspase_cys_AS
IPR001875 DED_dom
IPR016129 Caspase_his_AS
IPR040236 TMEM198
IPR025256 DUF4203
IPR001309 Pept_C14_p20
IPR015917 Pept_C14A
IPR002138 Pept_C14_p10
IPR002398 Pept_C14
IPR018484 Carb_kinase_FGGY_N
IPR018485 Carb_kinase_FGGY_C
IPR006003 FGGY_RbtK-like
IPR000352 Pep_chain_release_fac_I_II
IPR033139 Caspase_cys_AS
IPR001875 DED_dom
IPR016129 Caspase_his_AS
IPR040236 TMEM198
IPR025256 DUF4203
SUPFAM
SSF52129
SSF52129
ProteinModelPortal
H9JB31
A9ZMY8
A0A2A4KAV9
G0XQH8
G0XQH7
I7CAH9
+ More
A0A142J2X8 A0A0L7LPH4 R4I3K4 G0XQH9 F6K5S2 A0A194QL32 A0A1E1WQP3 A0A212FDA5 A0A194QJI1 G0XQH6 A0A346RAD7 D6WJH5 A0A3Q8C047 V5GYQ4 A0A3F2Z153 A0A1W4W5I4 A0A1W4WH04 A0A1W4W5J4 A0A182N2I9 A0A182HGZ6 A0A182L4D4 A0A182QBD6 A0A182VMT7 Q16H55 A0A1S4H7C5 W8AV04 A0A3F2YVF2 A3QVU6 A0A067QV28 A0A182SF59 W5JJK4 A0A084WBG7 A0A3F2YXQ1 A0A1B0D025 A0A182FTB8 A0A3F2YU15 A0A3F2YWL0 Q29IM7 A0A0R3NZ05 A0A0R3NYQ6 A0A1B0GLN8 B4M7Y9 A0A1I8M1X4 A0A0D3QBR4 B4N271 A0A0D3QC48 A0A0D3QBU5 A0A0D3QBU2 A0A0D3QBS0 A0A0D3QBQ6 A0A0D3QBR1 A0A1A9WRC9 A0A0D3QBR0 A0A0A1XM78 A0A0D3QC59 A0A0D3QBR6 A0A1Y1MQM3 A0A182H1W5 B4PX19 A0A1I8PI09 A0A0D3QCD4 A0A0M4EZM9 N6TGN6 B3P987 A0A0K8TMN0 B4JNA9 Q8IRY7 X2JHT0 A0A0L0CNR9 A0A336MAB9 A0A3Q9HGP0 A0A1B6JA75 B4H2Q9 A0A1B6HSW3 A0A0K1DH04 A0A1B0ALP5
A0A142J2X8 A0A0L7LPH4 R4I3K4 G0XQH9 F6K5S2 A0A194QL32 A0A1E1WQP3 A0A212FDA5 A0A194QJI1 G0XQH6 A0A346RAD7 D6WJH5 A0A3Q8C047 V5GYQ4 A0A3F2Z153 A0A1W4W5I4 A0A1W4WH04 A0A1W4W5J4 A0A182N2I9 A0A182HGZ6 A0A182L4D4 A0A182QBD6 A0A182VMT7 Q16H55 A0A1S4H7C5 W8AV04 A0A3F2YVF2 A3QVU6 A0A067QV28 A0A182SF59 W5JJK4 A0A084WBG7 A0A3F2YXQ1 A0A1B0D025 A0A182FTB8 A0A3F2YU15 A0A3F2YWL0 Q29IM7 A0A0R3NZ05 A0A0R3NYQ6 A0A1B0GLN8 B4M7Y9 A0A1I8M1X4 A0A0D3QBR4 B4N271 A0A0D3QC48 A0A0D3QBU5 A0A0D3QBU2 A0A0D3QBS0 A0A0D3QBQ6 A0A0D3QBR1 A0A1A9WRC9 A0A0D3QBR0 A0A0A1XM78 A0A0D3QC59 A0A0D3QBR6 A0A1Y1MQM3 A0A182H1W5 B4PX19 A0A1I8PI09 A0A0D3QCD4 A0A0M4EZM9 N6TGN6 B3P987 A0A0K8TMN0 B4JNA9 Q8IRY7 X2JHT0 A0A0L0CNR9 A0A336MAB9 A0A3Q9HGP0 A0A1B6JA75 B4H2Q9 A0A1B6HSW3 A0A0K1DH04 A0A1B0ALP5
PDB
4HVA
E-value=1.09895e-18,
Score=231
Ontologies
PATHWAY
GO
GO:0004197
GO:0006915
GO:0016301
GO:0016773
GO:0005975
GO:0003747
GO:0005737
GO:0097199
GO:0006919
GO:0097194
GO:0097153
GO:0016021
GO:0045087
GO:0050829
GO:0008656
GO:0006964
GO:0002230
GO:0061057
GO:0048935
GO:0043069
GO:0016485
GO:0007291
GO:0006955
GO:0045089
GO:0005829
GO:0006952
GO:0004175
GO:0006963
GO:0097200
GO:0008234
GO:0006508
GO:0004707
GO:0005524
GO:0031683
GO:0001664
GO:0005525
GO:0007264
GO:0003824
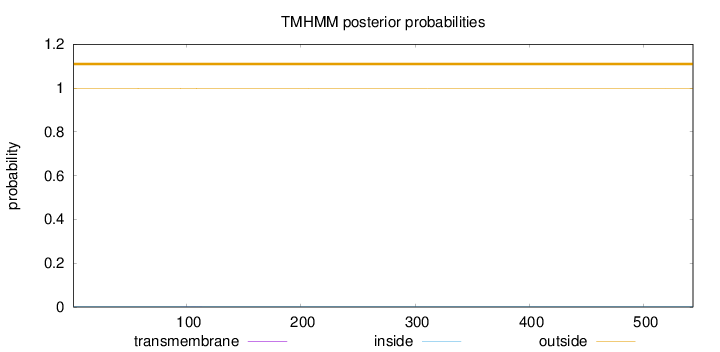
Topology
Subcellular location
Cytoplasm
Length:
543
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00780999999999998
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00129
outside
1 - 543
Population Genetic Test Statistics
Pi
120.349182
Theta
136.405243
Tajima's D
-0.807363
CLR
0.445834
CSRT
0.173341332933353
Interpretation
Uncertain
