Gene
KWMTBOMO05513
Pre Gene Modal
BGIBMGA006615
Annotation
PREDICTED:_probable_peptide_chain_release_factor_C12orf65_homolog?_mitochondrial_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.451
Sequence
CDS
ATGTTCTTGCTCACTTCAAAATATTTTAGAAATTTCAGTCCACGCTTAGCAGCTGCCTTTAAACATACAATAGATTATTCCAAAGTACCAAAGATCAATGAGTGTGAATTATCTGAAAAATTTGTTAGAGGAAGTGGCCCAGGAGGGTCAGCTGTAAACAAGAATTCTAATTGTGTGGTTCTTACACACATCCCCACAGGTACCGTCATAAAATGTCACACAAGTCGTTGTCAAGACGATAATCGGAAGAAAGCCCGGGAATTGTTAATTGAAAAACTGGATGATATGATCAATGGCCCTGAAAGTGTATCGGCGCAGAGGAAACTTATCGAAGAAAAAAAGTATAAACGGAATGCAACAAAAAAACAGAAAATATCCAAATTAAAAGATGAATGGAAGAAACGGGAAGGCATAATATAA
Protein
MFLLTSKYFRNFSPRLAAAFKHTIDYSKVPKINECELSEKFVRGSGPGGSAVNKNSNCVVLTHIPTGTVIKCHTSRCQDDNRKKARELLIEKLDDMINGPESVSAQRKLIEEKKYKRNATKKQKISKLKDEWKKREGII
Summary
Similarity
Belongs to the complex I LYR family.
Belongs to the peptidase C14A family.
Belongs to the peptidase C14A family.
Uniprot
H9JAS0
A0A2A4KA91
A0A212FD70
A0A2H1WUA7
A0A194QMU7
S4P6E3
+ More
A0A3S2NNL5 A0A0L7KWJ4 A0A194QJI1 A0A139WBI1 A0A1S3CUU9 A0A2S2P9I4 A0A1Y1M503 A0A2H8TLV1 A0A1B6IL58 A0A067QJ88 A0A2J7QIA8 A0A1B6EMI9 A0A1B6GAV5 J9K690 A0A1B6FAU9 A0A2P8YWX5 E9IC99 A0A1B0G2H6 A0A1B0ASP0 A0A1A9X5Y6 T1JD56 U5ERN3 W8AS58 F4W6J7 A0A034VHC2 A0A195D4D5 A0A195FIW3 A0A023EG47 A0A336M716 A0A1Q3FDI5 A0A2M4C2N3 A0A0J7KVU4 A0A1B0CIX3 A0A2M4C211 T1E7G6 A0A2M4C1I5 A0A195BT32 A0A2M4C1R4 A0A2M4C1S1 A0A131Y4F9 B0WTD3 A0A1Q3FDF9 A0A1Q3FCJ5 A0A1Q3FFY6 A0A1B6MJR7 A0A147BVD8 G3MTT4 A0A023G3R3 A0A158P2A3 A0A023G557 A0A1B6CSW7 Q16IE2 A0A023EE42 A0A023EE61 A0A224XQ80 A0A1Q3FGQ4 A0A0P4VH78 A0A1B0DP52 E2AU99 A0A2M3ZDK7 A0A161MH91 A0A0A9WA79 W5JGE5 Q291R9 A0A146MDV0 A0A0V0GBW1 E2C027 A0A2L2Y142 A0A293MUK3 B4GAC8 A0A182FKM5 A0A1I8NFA6 A0A1I8PVY9 A0A1L8E0D9 A0A0R1DY86 A0A195E461 A0A023EGH2 A0A182SHR9 A0A182NZ98 A0A0C9SAH2 A0A224Z8J5 A1ZAD7 A0A023FHE1 J3JUQ8 N6ULT0 B4HSV8 L7LYY0 A0A182IVG9 B3MFM5 A0A1L8E0B0 A0A1W4UZI5 A0A151X265 B4NVC0 A0A182WR41
A0A3S2NNL5 A0A0L7KWJ4 A0A194QJI1 A0A139WBI1 A0A1S3CUU9 A0A2S2P9I4 A0A1Y1M503 A0A2H8TLV1 A0A1B6IL58 A0A067QJ88 A0A2J7QIA8 A0A1B6EMI9 A0A1B6GAV5 J9K690 A0A1B6FAU9 A0A2P8YWX5 E9IC99 A0A1B0G2H6 A0A1B0ASP0 A0A1A9X5Y6 T1JD56 U5ERN3 W8AS58 F4W6J7 A0A034VHC2 A0A195D4D5 A0A195FIW3 A0A023EG47 A0A336M716 A0A1Q3FDI5 A0A2M4C2N3 A0A0J7KVU4 A0A1B0CIX3 A0A2M4C211 T1E7G6 A0A2M4C1I5 A0A195BT32 A0A2M4C1R4 A0A2M4C1S1 A0A131Y4F9 B0WTD3 A0A1Q3FDF9 A0A1Q3FCJ5 A0A1Q3FFY6 A0A1B6MJR7 A0A147BVD8 G3MTT4 A0A023G3R3 A0A158P2A3 A0A023G557 A0A1B6CSW7 Q16IE2 A0A023EE42 A0A023EE61 A0A224XQ80 A0A1Q3FGQ4 A0A0P4VH78 A0A1B0DP52 E2AU99 A0A2M3ZDK7 A0A161MH91 A0A0A9WA79 W5JGE5 Q291R9 A0A146MDV0 A0A0V0GBW1 E2C027 A0A2L2Y142 A0A293MUK3 B4GAC8 A0A182FKM5 A0A1I8NFA6 A0A1I8PVY9 A0A1L8E0D9 A0A0R1DY86 A0A195E461 A0A023EGH2 A0A182SHR9 A0A182NZ98 A0A0C9SAH2 A0A224Z8J5 A1ZAD7 A0A023FHE1 J3JUQ8 N6ULT0 B4HSV8 L7LYY0 A0A182IVG9 B3MFM5 A0A1L8E0B0 A0A1W4UZI5 A0A151X265 B4NVC0 A0A182WR41
Pubmed
19121390
22118469
26354079
23622113
26227816
18362917
+ More
19820115 28004739 24845553 29403074 21282665 24495485 21719571 25348373 24945155 29652888 22216098 21347285 17510324 27129103 20798317 25401762 20920257 23761445 15632085 17994087 26823975 26561354 25315136 17550304 26131772 28797301 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 23537049 25576852 22936249
19820115 28004739 24845553 29403074 21282665 24495485 21719571 25348373 24945155 29652888 22216098 21347285 17510324 27129103 20798317 25401762 20920257 23761445 15632085 17994087 26823975 26561354 25315136 17550304 26131772 28797301 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 23537049 25576852 22936249
EMBL
BABH01008520
NWSH01000001
PCG81127.1
AGBW02009100
OWR51689.1
ODYU01011114
+ More
SOQ56649.1 KQ461198 KPJ06285.1 GAIX01006876 JAA85684.1 RSAL01000201 RVE44433.1 JTDY01005001 KOB67490.1 KQ458793 KPJ05070.1 KQ971372 KYB25211.1 GGMR01013502 MBY26121.1 GEZM01040309 JAV80899.1 GFXV01003204 MBW15009.1 GECU01020046 GECU01007126 JAS87660.1 JAT00581.1 KK853300 KDR08762.1 NEVH01013957 PNF28309.1 GECZ01030658 JAS39111.1 GECZ01010208 JAS59561.1 ABLF02029696 GECZ01022518 JAS47251.1 PYGN01000312 PSN48735.1 GL762231 EFZ21809.1 CCAG010018314 JXJN01002890 JH432094 GANO01003667 JAB56204.1 GAMC01017648 JAB88907.1 GL887707 EGI70332.1 GAKP01017792 JAC41160.1 KQ976885 KYN07294.1 KQ981523 KYN40311.1 GAPW01005803 JAC07795.1 UFQS01000640 UFQT01000640 SSX05646.1 SSX26005.1 GFDL01009428 JAV25617.1 GGFJ01010117 MBW59258.1 LBMM01002773 KMQ94384.1 AJWK01013857 GGFJ01010050 MBW59191.1 GAMD01003072 JAA98518.1 GGFJ01010045 MBW59186.1 KQ976408 KYM91128.1 GGFJ01010119 MBW59260.1 GGFJ01010116 MBW59257.1 GEFM01002408 JAP73388.1 DS232085 EDS34371.1 GFDL01009434 JAV25611.1 GFDL01009741 JAV25304.1 GFDL01008582 JAV26463.1 GEBQ01003878 JAT36099.1 GEGO01000902 JAR94502.1 JO845286 AEO36902.1 GBBM01006517 JAC28901.1 ADTU01007045 GBBM01006516 JAC28902.1 GEDC01020738 JAS16560.1 CH478086 CH477828 EAT34024.1 EAT35857.1 GAPW01005996 JAC07602.1 GAPW01005700 JAC07898.1 GFTR01001770 JAW14656.1 GFDL01008307 JAV26738.1 GDKW01003696 JAI52899.1 AJVK01018086 GL442815 EFN62956.1 GGFM01005841 MBW26592.1 GEMB01002500 JAS00686.1 GBHO01038940 GBRD01010572 GBRD01001890 JAG04664.1 JAG55252.1 ADMH02001585 ETN61965.1 CM000071 EAL25043.3 GDHC01001437 JAQ17192.1 GECL01000739 JAP05385.1 GL451712 EFN78723.1 IAAA01007888 LAA01883.1 GFWV01019754 MAA44482.1 CH479181 EDW31880.1 GFDF01001968 JAV12116.1 CM000158 KRK00218.1 KQ979657 KYN19943.1 GAPW01005804 JAC07794.1 GBZX01002588 JAG90152.1 GFPF01012755 MAA23901.1 AE013599 BT030757 AAM68513.1 ABV82139.1 AHN56281.1 GBBK01004043 JAC20439.1 BT126973 AEE61935.1 APGK01018751 KB740082 KB631720 ENN81596.1 ERL85577.1 CH480816 EDW48122.1 GACK01008029 JAA57005.1 CH902619 EDV37715.1 GFDF01001967 JAV12117.1 KQ982580 KYQ54497.1 CH987229 CM002911 EDX16001.1 KMY94236.1
SOQ56649.1 KQ461198 KPJ06285.1 GAIX01006876 JAA85684.1 RSAL01000201 RVE44433.1 JTDY01005001 KOB67490.1 KQ458793 KPJ05070.1 KQ971372 KYB25211.1 GGMR01013502 MBY26121.1 GEZM01040309 JAV80899.1 GFXV01003204 MBW15009.1 GECU01020046 GECU01007126 JAS87660.1 JAT00581.1 KK853300 KDR08762.1 NEVH01013957 PNF28309.1 GECZ01030658 JAS39111.1 GECZ01010208 JAS59561.1 ABLF02029696 GECZ01022518 JAS47251.1 PYGN01000312 PSN48735.1 GL762231 EFZ21809.1 CCAG010018314 JXJN01002890 JH432094 GANO01003667 JAB56204.1 GAMC01017648 JAB88907.1 GL887707 EGI70332.1 GAKP01017792 JAC41160.1 KQ976885 KYN07294.1 KQ981523 KYN40311.1 GAPW01005803 JAC07795.1 UFQS01000640 UFQT01000640 SSX05646.1 SSX26005.1 GFDL01009428 JAV25617.1 GGFJ01010117 MBW59258.1 LBMM01002773 KMQ94384.1 AJWK01013857 GGFJ01010050 MBW59191.1 GAMD01003072 JAA98518.1 GGFJ01010045 MBW59186.1 KQ976408 KYM91128.1 GGFJ01010119 MBW59260.1 GGFJ01010116 MBW59257.1 GEFM01002408 JAP73388.1 DS232085 EDS34371.1 GFDL01009434 JAV25611.1 GFDL01009741 JAV25304.1 GFDL01008582 JAV26463.1 GEBQ01003878 JAT36099.1 GEGO01000902 JAR94502.1 JO845286 AEO36902.1 GBBM01006517 JAC28901.1 ADTU01007045 GBBM01006516 JAC28902.1 GEDC01020738 JAS16560.1 CH478086 CH477828 EAT34024.1 EAT35857.1 GAPW01005996 JAC07602.1 GAPW01005700 JAC07898.1 GFTR01001770 JAW14656.1 GFDL01008307 JAV26738.1 GDKW01003696 JAI52899.1 AJVK01018086 GL442815 EFN62956.1 GGFM01005841 MBW26592.1 GEMB01002500 JAS00686.1 GBHO01038940 GBRD01010572 GBRD01001890 JAG04664.1 JAG55252.1 ADMH02001585 ETN61965.1 CM000071 EAL25043.3 GDHC01001437 JAQ17192.1 GECL01000739 JAP05385.1 GL451712 EFN78723.1 IAAA01007888 LAA01883.1 GFWV01019754 MAA44482.1 CH479181 EDW31880.1 GFDF01001968 JAV12116.1 CM000158 KRK00218.1 KQ979657 KYN19943.1 GAPW01005804 JAC07794.1 GBZX01002588 JAG90152.1 GFPF01012755 MAA23901.1 AE013599 BT030757 AAM68513.1 ABV82139.1 AHN56281.1 GBBK01004043 JAC20439.1 BT126973 AEE61935.1 APGK01018751 KB740082 KB631720 ENN81596.1 ERL85577.1 CH480816 EDW48122.1 GACK01008029 JAA57005.1 CH902619 EDV37715.1 GFDF01001967 JAV12117.1 KQ982580 KYQ54497.1 CH987229 CM002911 EDX16001.1 KMY94236.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000283053
UP000037510
+ More
UP000053268 UP000007266 UP000079169 UP000027135 UP000235965 UP000007819 UP000245037 UP000092444 UP000092460 UP000092443 UP000007755 UP000078542 UP000078541 UP000036403 UP000092461 UP000078540 UP000002320 UP000005205 UP000008820 UP000092462 UP000000311 UP000000673 UP000001819 UP000008237 UP000008744 UP000069272 UP000095301 UP000095300 UP000002282 UP000078492 UP000075901 UP000075884 UP000000803 UP000019118 UP000030742 UP000001292 UP000075880 UP000007801 UP000192221 UP000075809 UP000000304 UP000075920
UP000053268 UP000007266 UP000079169 UP000027135 UP000235965 UP000007819 UP000245037 UP000092444 UP000092460 UP000092443 UP000007755 UP000078542 UP000078541 UP000036403 UP000092461 UP000078540 UP000002320 UP000005205 UP000008820 UP000092462 UP000000311 UP000000673 UP000001819 UP000008237 UP000008744 UP000069272 UP000095301 UP000095300 UP000002282 UP000078492 UP000075901 UP000075884 UP000000803 UP000019118 UP000030742 UP000001292 UP000075880 UP000007801 UP000192221 UP000075809 UP000000304 UP000075920
Interpro
SUPFAM
SSF52129
SSF52129
ProteinModelPortal
H9JAS0
A0A2A4KA91
A0A212FD70
A0A2H1WUA7
A0A194QMU7
S4P6E3
+ More
A0A3S2NNL5 A0A0L7KWJ4 A0A194QJI1 A0A139WBI1 A0A1S3CUU9 A0A2S2P9I4 A0A1Y1M503 A0A2H8TLV1 A0A1B6IL58 A0A067QJ88 A0A2J7QIA8 A0A1B6EMI9 A0A1B6GAV5 J9K690 A0A1B6FAU9 A0A2P8YWX5 E9IC99 A0A1B0G2H6 A0A1B0ASP0 A0A1A9X5Y6 T1JD56 U5ERN3 W8AS58 F4W6J7 A0A034VHC2 A0A195D4D5 A0A195FIW3 A0A023EG47 A0A336M716 A0A1Q3FDI5 A0A2M4C2N3 A0A0J7KVU4 A0A1B0CIX3 A0A2M4C211 T1E7G6 A0A2M4C1I5 A0A195BT32 A0A2M4C1R4 A0A2M4C1S1 A0A131Y4F9 B0WTD3 A0A1Q3FDF9 A0A1Q3FCJ5 A0A1Q3FFY6 A0A1B6MJR7 A0A147BVD8 G3MTT4 A0A023G3R3 A0A158P2A3 A0A023G557 A0A1B6CSW7 Q16IE2 A0A023EE42 A0A023EE61 A0A224XQ80 A0A1Q3FGQ4 A0A0P4VH78 A0A1B0DP52 E2AU99 A0A2M3ZDK7 A0A161MH91 A0A0A9WA79 W5JGE5 Q291R9 A0A146MDV0 A0A0V0GBW1 E2C027 A0A2L2Y142 A0A293MUK3 B4GAC8 A0A182FKM5 A0A1I8NFA6 A0A1I8PVY9 A0A1L8E0D9 A0A0R1DY86 A0A195E461 A0A023EGH2 A0A182SHR9 A0A182NZ98 A0A0C9SAH2 A0A224Z8J5 A1ZAD7 A0A023FHE1 J3JUQ8 N6ULT0 B4HSV8 L7LYY0 A0A182IVG9 B3MFM5 A0A1L8E0B0 A0A1W4UZI5 A0A151X265 B4NVC0 A0A182WR41
A0A3S2NNL5 A0A0L7KWJ4 A0A194QJI1 A0A139WBI1 A0A1S3CUU9 A0A2S2P9I4 A0A1Y1M503 A0A2H8TLV1 A0A1B6IL58 A0A067QJ88 A0A2J7QIA8 A0A1B6EMI9 A0A1B6GAV5 J9K690 A0A1B6FAU9 A0A2P8YWX5 E9IC99 A0A1B0G2H6 A0A1B0ASP0 A0A1A9X5Y6 T1JD56 U5ERN3 W8AS58 F4W6J7 A0A034VHC2 A0A195D4D5 A0A195FIW3 A0A023EG47 A0A336M716 A0A1Q3FDI5 A0A2M4C2N3 A0A0J7KVU4 A0A1B0CIX3 A0A2M4C211 T1E7G6 A0A2M4C1I5 A0A195BT32 A0A2M4C1R4 A0A2M4C1S1 A0A131Y4F9 B0WTD3 A0A1Q3FDF9 A0A1Q3FCJ5 A0A1Q3FFY6 A0A1B6MJR7 A0A147BVD8 G3MTT4 A0A023G3R3 A0A158P2A3 A0A023G557 A0A1B6CSW7 Q16IE2 A0A023EE42 A0A023EE61 A0A224XQ80 A0A1Q3FGQ4 A0A0P4VH78 A0A1B0DP52 E2AU99 A0A2M3ZDK7 A0A161MH91 A0A0A9WA79 W5JGE5 Q291R9 A0A146MDV0 A0A0V0GBW1 E2C027 A0A2L2Y142 A0A293MUK3 B4GAC8 A0A182FKM5 A0A1I8NFA6 A0A1I8PVY9 A0A1L8E0D9 A0A0R1DY86 A0A195E461 A0A023EGH2 A0A182SHR9 A0A182NZ98 A0A0C9SAH2 A0A224Z8J5 A1ZAD7 A0A023FHE1 J3JUQ8 N6ULT0 B4HSV8 L7LYY0 A0A182IVG9 B3MFM5 A0A1L8E0B0 A0A1W4UZI5 A0A151X265 B4NVC0 A0A182WR41
PDB
2RSM
E-value=1.00516e-19,
Score=231
Ontologies
GO
PANTHER
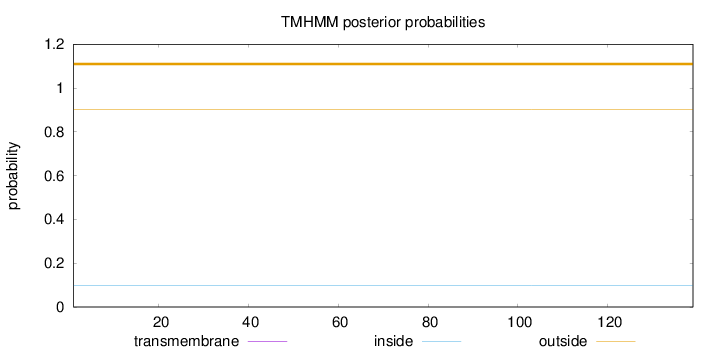
Topology
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00053
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.09696
outside
1 - 139
Population Genetic Test Statistics
Pi
165.931017
Theta
139.24408
Tajima's D
0.37847
CLR
228.442804
CSRT
0.475376231188441
Interpretation
Uncertain
