Pre Gene Modal
BGIBMGA006725
Annotation
extracellular_regulated_MAP_kinase_[Bombyx_mori]
Full name
Mitogen-activated protein kinase
Location in the cell
Cytoplasmic Reliability : 2.219
Sequence
CDS
ATGAAAGACGTTTACATCGTTCAATGCTTGATGGAGACCGATCTTTACAAGCTGTTGAAGACCCAGAAATTAAGTAACGATCACATCTGCTACTTCCTGTATCAGATCCTTCGAGGACTGAAATATATTCACTCTGCAAACGTCCTCCATAGGGACTTGAAGCCTTCTAATTTACTACTTAATACCACGTGCGATTTGAAGATTTGCGACTTCGGTCTGGCGCGAGTTGCGGATCCTGATCACGACCACACCGGTTTCCTCACGGAATACGTAGCCACGCGCTGGTACCGTGCTCCTGAAATCATGCTCAACTCAAAGGGCTATACCAAATCAATAGATATTTGGTCAGTGGGATGTATTTTGGCGGAGATGCTGTCCAACAGACCTATATTCCCGGGCAAGCACTATTTGGATCAATTGAATCACATTTTGGGAGTGCTCGGTTCGCCGAGTCAGGAGGATCTTGATTGCATCATTAACGAAAAGGCTCGTGGTTACTTGGAGTCGTTGCCGTTCAAGCCTCGCGTGCCGTGGGTGGACCTCTTCCCCGGAGCGGACCCTCGCGCCTTGGACCTGCTCCATCGCATGCTGACTTTCAACCCTCACAAACGCATCACGGTCGAGGAAGCCCTAGCCCATCCGTACCTCGAGCAGTACTACGACCCGAATGATGAGCCCGTAGCCGAGGAGCCTTTCCGTTTCGCGATGGAGCTCGACGATCTCCCGAAGGAGACGCTCAAGAAATACATATTCGAGGAGACGTTGTTCTTTAAACAGCAGAACGAAGACGCGTAA
Protein
MKDVYIVQCLMETDLYKLLKTQKLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTTCDLKICDFGLARVADPDHDHTGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLSNRPIFPGKHYLDQLNHILGVLGSPSQEDLDCIINEKARGYLESLPFKPRVPWVDLFPGADPRALDLLHRMLTFNPHKRITVEEALAHPYLEQYYDPNDEPVAEEPFRFAMELDDLPKETLKKYIFEETLFFKQQNEDA
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. MAP kinase subfamily.
Uniprot
Q3C2L4
H9JB30
A0A2H1WDP4
A0A194QI47
A0A345CI05
A0A2A4J871
+ More
A0A1B0YY60 A0A1Y1K5A3 A0A1Y1K4F9 A0A1B6LW59 A0A2J7QM98 A0A1B6GNA8 R4WCW8 A0A0L7LF64 A0A1B6DKW0 A0A067RFT2 V5I8E7 A0A1I9WLG8 U5EVN2 A0A224XR06 A0A023FCL1 A0A069DYR7 S5TBX9 K9LM34 G8Z9H6 V5N6T9 A0A0A9WY65 A0A1J1I3C1 A0A0P6IXU9 T1IFJ4 A0A0P4VKF7 A0A182QR63 B0WDB3 G3GC05 Q16HR0 A0A1Q3G0K2 J9PCW7 A0A1A9W5C4 T1E242 C6L430 A7BHS4 A0A0P4W996 D0EKI4 A0A182IZZ6 A0A0M4M1D3 Q7PVR1 A0A1S4H175 A0A182I592 A0A2M4CWD2 A0A2M4CV60 A0A2M4AI56 A0A2M3ZKR3 A0A0L0CBJ2 E2BZZ2 A0A0K8TW32 A0A2K6VB43 T1E1K2 A0A1S3J4P5 A0A182Y9U8 A0A182RG65 A0A232F3U0 A0A034W3U1 A0A195ERA1 F4W648 A0A158P1W9 A0A195DMU6 A0A151I7W4 A0A0L7RJA8 A0A0U2DB09 A0A195AWA8 A0A154PD45 A0A026WAQ7 T1PAX6 A0A0A1X1V3 A0A0P5PHU9 A0A3L8DR75 Q16945 A0A0P5BRV5 A0A1B0A4T4 A0A3S3QDC1 J7FZ52 E1ZZM3 A0A293L4K2 A0A1B0G2A4 A0A1B0ANY6 A0A1A9XVU3 A0A1A9V2A9 D6X2Z2 A0A1L7H9J9 A0A2C9K4U6 A0A1I8P312 A0A2R5L8W0 A0A310SAP0 A0A0N8BWE0 R7V580 A0A151X0J9 A0A0M8ZPJ6 A0A1L6UW76 A0A2S2P210 A0A0P6JQZ7
A0A1B0YY60 A0A1Y1K5A3 A0A1Y1K4F9 A0A1B6LW59 A0A2J7QM98 A0A1B6GNA8 R4WCW8 A0A0L7LF64 A0A1B6DKW0 A0A067RFT2 V5I8E7 A0A1I9WLG8 U5EVN2 A0A224XR06 A0A023FCL1 A0A069DYR7 S5TBX9 K9LM34 G8Z9H6 V5N6T9 A0A0A9WY65 A0A1J1I3C1 A0A0P6IXU9 T1IFJ4 A0A0P4VKF7 A0A182QR63 B0WDB3 G3GC05 Q16HR0 A0A1Q3G0K2 J9PCW7 A0A1A9W5C4 T1E242 C6L430 A7BHS4 A0A0P4W996 D0EKI4 A0A182IZZ6 A0A0M4M1D3 Q7PVR1 A0A1S4H175 A0A182I592 A0A2M4CWD2 A0A2M4CV60 A0A2M4AI56 A0A2M3ZKR3 A0A0L0CBJ2 E2BZZ2 A0A0K8TW32 A0A2K6VB43 T1E1K2 A0A1S3J4P5 A0A182Y9U8 A0A182RG65 A0A232F3U0 A0A034W3U1 A0A195ERA1 F4W648 A0A158P1W9 A0A195DMU6 A0A151I7W4 A0A0L7RJA8 A0A0U2DB09 A0A195AWA8 A0A154PD45 A0A026WAQ7 T1PAX6 A0A0A1X1V3 A0A0P5PHU9 A0A3L8DR75 Q16945 A0A0P5BRV5 A0A1B0A4T4 A0A3S3QDC1 J7FZ52 E1ZZM3 A0A293L4K2 A0A1B0G2A4 A0A1B0ANY6 A0A1A9XVU3 A0A1A9V2A9 D6X2Z2 A0A1L7H9J9 A0A2C9K4U6 A0A1I8P312 A0A2R5L8W0 A0A310SAP0 A0A0N8BWE0 R7V580 A0A151X0J9 A0A0M8ZPJ6 A0A1L6UW76 A0A2S2P210 A0A0P6JQZ7
EC Number
2.7.11.24
Pubmed
16360949
19121390
26354079
28004739
23691247
26227816
+ More
24845553 27538518 25474469 26334808 24084041 25401762 26823975 26999592 27129103 17510324 24330624 26846853 17681525 22394010 12364791 26108605 20798317 20920257 25244985 28648823 25348373 21719571 21347285 24508170 25315136 25830018 30249741 18362917 19820115 27765698 15562597 23254933 28012735
24845553 27538518 25474469 26334808 24084041 25401762 26823975 26999592 27129103 17510324 24330624 26846853 17681525 22394010 12364791 26108605 20798317 20920257 25244985 28648823 25348373 21719571 21347285 24508170 25315136 25830018 30249741 18362917 19820115 27765698 15562597 23254933 28012735
EMBL
AB183869
BAE46741.1
BABH01008514
ODYU01007854
SOQ50992.1
KQ458793
+ More
KPJ05069.1 MF797866 AXF67444.1 NWSH01002739 PCG67602.1 KU358552 RSAL01000201 ANO46362.1 RVE44434.1 GEZM01095789 JAV55350.1 GEZM01095790 JAV55348.1 GEBQ01012066 JAT27911.1 NEVH01013214 PNF29720.1 GECZ01015572 GECZ01005862 JAS54197.1 JAS63907.1 AK417233 BAN20448.1 JTDY01001350 KOB74107.1 GEDC01027332 GEDC01011048 JAS09966.1 JAS26250.1 KK852530 KDR21908.1 GALX01004815 JAB63651.1 KU932352 APA33988.1 GANO01003332 JAB56539.1 GFTR01005434 JAW10992.1 GBBI01000179 JAC18533.1 GBGD01001920 JAC86969.1 KC896761 AGS38337.1 JN035901 AFL70596.1 GU324353 GU324354 ADT80930.1 KC537791 AHA83424.1 GBHO01030187 GDHC01007988 JAG13417.1 JAQ10641.1 CVRI01000038 CRK94274.1 GDUN01000123 JAN95796.1 ACPB03005609 GDKW01001785 JAI54810.1 AXCN02000825 DS231895 EDS44348.1 JF830802 AEE81046.1 CH478148 EAT33794.1 GFDL01001728 JAV33317.1 GU002542 KP100030 ADF87942.1 AKO62680.1 GALA01001074 JAA93778.1 KU160504 AB478860 AME17868.1 BAH86598.1 AB277827 BAF75365.1 GDRN01076339 JAI62909.1 GQ847862 ACX32460.1 KR075869 ALE20588.1 AAAB01008984 EAA14714.5 APCN01003837 GGFL01004980 MBW69158.1 GGFL01004981 MBW69159.1 GGFK01007154 MBW40475.1 GGFM01008373 MBW29124.1 JRES01000648 KNC29602.1 GL451708 EFN78769.1 GDHF01034054 JAI18260.1 GALA01001589 JAA93263.1 NNAY01001066 OXU25262.1 GAKP01009568 JAC49384.1 KQ982021 KYN30409.1 GL887707 EGI70183.1 ADTU01006906 KQ980724 KYN14146.1 KQ978397 KYM94213.1 KQ414582 KOC70909.1 KP099006 AKO62652.1 KQ976731 KYM76320.1 KQ434874 KZC09743.1 KK107293 EZA53172.1 KA645305 AFP59934.1 GBXI01009185 JAD05107.1 GDIQ01138340 JAL13386.1 QOIP01000005 RLU22389.1 U40484 AAA83210.1 GDIP01181690 GDIQ01123541 JAJ41712.1 JAL28185.1 NCKU01003464 RWS07466.1 JX103496 AFP57674.1 GL435444 EFN73366.1 GFWV01001097 MAA25827.1 CCAG010004344 JXJN01001079 KQ971372 EFA10708.1 KU351558 APU52030.1 GGLE01001784 MBY05910.1 KQ766110 OAD53741.1 GDIQ01138339 JAL13387.1 AMQN01000736 AMQN01000737 KB295062 ELU13684.1 KQ982611 KYQ53901.1 KQ435922 KOX68500.1 KY283808 APS85777.1 GGMR01010841 MBY23460.1 GDIQ01011806 JAN82931.1
KPJ05069.1 MF797866 AXF67444.1 NWSH01002739 PCG67602.1 KU358552 RSAL01000201 ANO46362.1 RVE44434.1 GEZM01095789 JAV55350.1 GEZM01095790 JAV55348.1 GEBQ01012066 JAT27911.1 NEVH01013214 PNF29720.1 GECZ01015572 GECZ01005862 JAS54197.1 JAS63907.1 AK417233 BAN20448.1 JTDY01001350 KOB74107.1 GEDC01027332 GEDC01011048 JAS09966.1 JAS26250.1 KK852530 KDR21908.1 GALX01004815 JAB63651.1 KU932352 APA33988.1 GANO01003332 JAB56539.1 GFTR01005434 JAW10992.1 GBBI01000179 JAC18533.1 GBGD01001920 JAC86969.1 KC896761 AGS38337.1 JN035901 AFL70596.1 GU324353 GU324354 ADT80930.1 KC537791 AHA83424.1 GBHO01030187 GDHC01007988 JAG13417.1 JAQ10641.1 CVRI01000038 CRK94274.1 GDUN01000123 JAN95796.1 ACPB03005609 GDKW01001785 JAI54810.1 AXCN02000825 DS231895 EDS44348.1 JF830802 AEE81046.1 CH478148 EAT33794.1 GFDL01001728 JAV33317.1 GU002542 KP100030 ADF87942.1 AKO62680.1 GALA01001074 JAA93778.1 KU160504 AB478860 AME17868.1 BAH86598.1 AB277827 BAF75365.1 GDRN01076339 JAI62909.1 GQ847862 ACX32460.1 KR075869 ALE20588.1 AAAB01008984 EAA14714.5 APCN01003837 GGFL01004980 MBW69158.1 GGFL01004981 MBW69159.1 GGFK01007154 MBW40475.1 GGFM01008373 MBW29124.1 JRES01000648 KNC29602.1 GL451708 EFN78769.1 GDHF01034054 JAI18260.1 GALA01001589 JAA93263.1 NNAY01001066 OXU25262.1 GAKP01009568 JAC49384.1 KQ982021 KYN30409.1 GL887707 EGI70183.1 ADTU01006906 KQ980724 KYN14146.1 KQ978397 KYM94213.1 KQ414582 KOC70909.1 KP099006 AKO62652.1 KQ976731 KYM76320.1 KQ434874 KZC09743.1 KK107293 EZA53172.1 KA645305 AFP59934.1 GBXI01009185 JAD05107.1 GDIQ01138340 JAL13386.1 QOIP01000005 RLU22389.1 U40484 AAA83210.1 GDIP01181690 GDIQ01123541 JAJ41712.1 JAL28185.1 NCKU01003464 RWS07466.1 JX103496 AFP57674.1 GL435444 EFN73366.1 GFWV01001097 MAA25827.1 CCAG010004344 JXJN01001079 KQ971372 EFA10708.1 KU351558 APU52030.1 GGLE01001784 MBY05910.1 KQ766110 OAD53741.1 GDIQ01138339 JAL13387.1 AMQN01000736 AMQN01000737 KB295062 ELU13684.1 KQ982611 KYQ53901.1 KQ435922 KOX68500.1 KY283808 APS85777.1 GGMR01010841 MBY23460.1 GDIQ01011806 JAN82931.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000283053
UP000235965
UP000037510
+ More
UP000027135 UP000183832 UP000015103 UP000075886 UP000002320 UP000008820 UP000091820 UP000075880 UP000007062 UP000075840 UP000037069 UP000008237 UP000085678 UP000076408 UP000075900 UP000215335 UP000078541 UP000007755 UP000005205 UP000078492 UP000078542 UP000053825 UP000078540 UP000076502 UP000053097 UP000095301 UP000279307 UP000092445 UP000285301 UP000000311 UP000092444 UP000092460 UP000092443 UP000078200 UP000007266 UP000076420 UP000095300 UP000014760 UP000075809 UP000053105
UP000027135 UP000183832 UP000015103 UP000075886 UP000002320 UP000008820 UP000091820 UP000075880 UP000007062 UP000075840 UP000037069 UP000008237 UP000085678 UP000076408 UP000075900 UP000215335 UP000078541 UP000007755 UP000005205 UP000078492 UP000078542 UP000053825 UP000078540 UP000076502 UP000053097 UP000095301 UP000279307 UP000092445 UP000285301 UP000000311 UP000092444 UP000092460 UP000092443 UP000078200 UP000007266 UP000076420 UP000095300 UP000014760 UP000075809 UP000053105
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Q3C2L4
H9JB30
A0A2H1WDP4
A0A194QI47
A0A345CI05
A0A2A4J871
+ More
A0A1B0YY60 A0A1Y1K5A3 A0A1Y1K4F9 A0A1B6LW59 A0A2J7QM98 A0A1B6GNA8 R4WCW8 A0A0L7LF64 A0A1B6DKW0 A0A067RFT2 V5I8E7 A0A1I9WLG8 U5EVN2 A0A224XR06 A0A023FCL1 A0A069DYR7 S5TBX9 K9LM34 G8Z9H6 V5N6T9 A0A0A9WY65 A0A1J1I3C1 A0A0P6IXU9 T1IFJ4 A0A0P4VKF7 A0A182QR63 B0WDB3 G3GC05 Q16HR0 A0A1Q3G0K2 J9PCW7 A0A1A9W5C4 T1E242 C6L430 A7BHS4 A0A0P4W996 D0EKI4 A0A182IZZ6 A0A0M4M1D3 Q7PVR1 A0A1S4H175 A0A182I592 A0A2M4CWD2 A0A2M4CV60 A0A2M4AI56 A0A2M3ZKR3 A0A0L0CBJ2 E2BZZ2 A0A0K8TW32 A0A2K6VB43 T1E1K2 A0A1S3J4P5 A0A182Y9U8 A0A182RG65 A0A232F3U0 A0A034W3U1 A0A195ERA1 F4W648 A0A158P1W9 A0A195DMU6 A0A151I7W4 A0A0L7RJA8 A0A0U2DB09 A0A195AWA8 A0A154PD45 A0A026WAQ7 T1PAX6 A0A0A1X1V3 A0A0P5PHU9 A0A3L8DR75 Q16945 A0A0P5BRV5 A0A1B0A4T4 A0A3S3QDC1 J7FZ52 E1ZZM3 A0A293L4K2 A0A1B0G2A4 A0A1B0ANY6 A0A1A9XVU3 A0A1A9V2A9 D6X2Z2 A0A1L7H9J9 A0A2C9K4U6 A0A1I8P312 A0A2R5L8W0 A0A310SAP0 A0A0N8BWE0 R7V580 A0A151X0J9 A0A0M8ZPJ6 A0A1L6UW76 A0A2S2P210 A0A0P6JQZ7
A0A1B0YY60 A0A1Y1K5A3 A0A1Y1K4F9 A0A1B6LW59 A0A2J7QM98 A0A1B6GNA8 R4WCW8 A0A0L7LF64 A0A1B6DKW0 A0A067RFT2 V5I8E7 A0A1I9WLG8 U5EVN2 A0A224XR06 A0A023FCL1 A0A069DYR7 S5TBX9 K9LM34 G8Z9H6 V5N6T9 A0A0A9WY65 A0A1J1I3C1 A0A0P6IXU9 T1IFJ4 A0A0P4VKF7 A0A182QR63 B0WDB3 G3GC05 Q16HR0 A0A1Q3G0K2 J9PCW7 A0A1A9W5C4 T1E242 C6L430 A7BHS4 A0A0P4W996 D0EKI4 A0A182IZZ6 A0A0M4M1D3 Q7PVR1 A0A1S4H175 A0A182I592 A0A2M4CWD2 A0A2M4CV60 A0A2M4AI56 A0A2M3ZKR3 A0A0L0CBJ2 E2BZZ2 A0A0K8TW32 A0A2K6VB43 T1E1K2 A0A1S3J4P5 A0A182Y9U8 A0A182RG65 A0A232F3U0 A0A034W3U1 A0A195ERA1 F4W648 A0A158P1W9 A0A195DMU6 A0A151I7W4 A0A0L7RJA8 A0A0U2DB09 A0A195AWA8 A0A154PD45 A0A026WAQ7 T1PAX6 A0A0A1X1V3 A0A0P5PHU9 A0A3L8DR75 Q16945 A0A0P5BRV5 A0A1B0A4T4 A0A3S3QDC1 J7FZ52 E1ZZM3 A0A293L4K2 A0A1B0G2A4 A0A1B0ANY6 A0A1A9XVU3 A0A1A9V2A9 D6X2Z2 A0A1L7H9J9 A0A2C9K4U6 A0A1I8P312 A0A2R5L8W0 A0A310SAP0 A0A0N8BWE0 R7V580 A0A151X0J9 A0A0M8ZPJ6 A0A1L6UW76 A0A2S2P210 A0A0P6JQZ7
PDB
2ZOQ
E-value=2.01899e-134,
Score=1225
Ontologies
PATHWAY
04013
MAPK signaling pathway - fly - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04320 Dorso-ventral axis formation - Bombyx mori (domestic silkworm)
04350 TGF-beta signaling pathway - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04214 Apoptosis - fly - Bombyx mori (domestic silkworm)
04320 Dorso-ventral axis formation - Bombyx mori (domestic silkworm)
04350 TGF-beta signaling pathway - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
GO
Topology
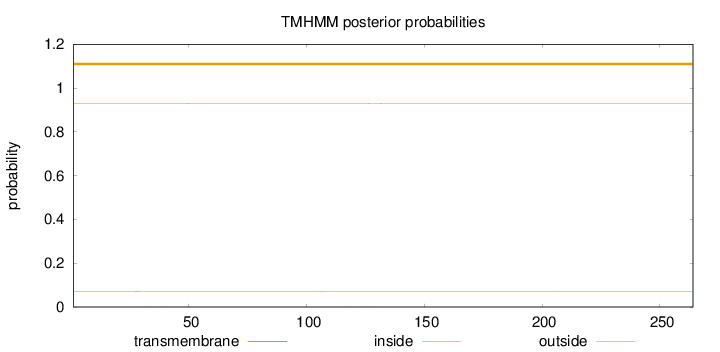
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02396
Exp number, first 60 AAs:
0.01119
Total prob of N-in:
0.07168
outside
1 - 264
Population Genetic Test Statistics
Pi
185.040927
Theta
183.70505
Tajima's D
0.027802
CLR
0.595672
CSRT
0.374031298435078
Interpretation
Uncertain
