Gene
KWMTBOMO05511
Pre Gene Modal
BGIBMGA006616
Annotation
ORAI_calcium_release-activated_calcium_modulator_1_isoform_B_[Bombyx_mori]
Full name
Calcium release-activated calcium channel protein 1
Alternative Name
Protein orai
dOrai
dOrai
Location in the cell
PlasmaMembrane Reliability : 4.77
Sequence
CDS
ATGTCGGGTGAGACGCCGATACAGTCGGGGGACGGGCTTCATACGCCGGCGTACCTCTCGTGGAGGAAGTTGCAGCTTAGTAGGGCGAAACTGAAGGCTTCCAGCAAAACATCTGCCCTACTTTCCGGTTTCGCTATGGTGGCTATGGTCGAAGTACAATTAAATCCAGCCCCCACTGCAGTACCAAAGGAGATGCTCGTCGCTTTCACGGTGTGCACAACACTCTTAGTCGCAGTCCACATGCTCGCGCTCATGATTAGCACCTGCATTTTGCCAAACATCGAAGCAGTCGGCAACTTGCATAGTATAGCGCTCGTCCATGAGTCGCCACACGAGAGACTCCACTGGTACATCGAAGTGGCTTGGGCTTTTTCGACCCTCTTAGGCTTAATCTTATTCTTGATAGAGATAGCTATACTATGTTGGGTTAAATTCTACGATTTGAGTCCGACGGCAGCTTGGTCGGCCTGTGTGGTCCTTATACCTGTTATGATTGTATTTTTGGCGTTCGCAATACATTTTTACATGTCCCTGGCGACGCACAAGTACGAAGTCACGGTGACAGGTATCAAAGAATTAGAATTGCTCAAAGAACAGATCGAAATGGGCGACCACGACGCCAGAATGAACAGCTTAACGTTGCTGGACCAAAGTCGGGTCGTCTGA
Protein
MSGETPIQSGDGLHTPAYLSWRKLQLSRAKLKASSKTSALLSGFAMVAMVEVQLNPAPTAVPKEMLVAFTVCTTLLVAVHMLALMISTCILPNIEAVGNLHSIALVHESPHERLHWYIEVAWAFSTLLGLILFLIEIAILCWVKFYDLSPTAAWSACVVLIPVMIVFLAFAIHFYMSLATHKYEVTVTGIKELELLKEQIEMGDHDARMNSLTLLDQSRVV
Summary
Description
Ca(2+) release-activated Ca(2+) (CRAC) channel subunit which mediates Ca(2+) influx following depletion of intracellular Ca(2+) stores. Regulates transcription factor NFAT nuclear import.
Miscellaneous
In Greek mythology, the 'Orai' are the keepers of the gates of heaven: Eunomia (order or harmony), Dike (justice) and Eirene (peace).
Similarity
Belongs to the Orai family.
Keywords
3D-structure
Alternative splicing
Calcium
Calcium channel
Calcium transport
Cell membrane
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Calcium release-activated calcium channel protein 1
splice variant In isoform E.
splice variant In isoform E.
Uniprot
B5BRC4
B5BRC5
A0A2A4J677
A0A2A4J5M0
A0A2H4THI5
A0A2A4J4V7
+ More
A0A2H1X1P9 A0A194RNY3 A0A194QHP9 A0A212FPV1 A0A0L7LFC4 A0A1E1W0N2 A0A2J7PD50 A0A1B6BWH1 A0A067RKQ5 A0A2A3E349 V9IMF7 A0A1B6DQT6 A0A1B6DJR3 A0A0C9S228 E0VCV9 A0A0Q9WFR7 E1ZXA4 B4KQU4 K7IRN4 A0A0N0BC83 A0A154PPF8 A0A0L7R8M8 A0A1B6DTE8 A0A195FT39 A0A195BD97 A0A195D4X5 A0A195DI95 A0A3L8E2D9 A0A026WLE9 B4LL00 A0A088A6F8 A0A158NQQ6 E2BPB3 A0A0Q9XLG5 F4WBV4 A0A151XB59 A0A0M4EFW8 A0A1J1IU05 E9IG63 A0A232F4J2 A0A023ENZ4 A0A182GIF4 A0A023EP20 Q1DGX6 A0A336MRH2 Q17KQ6 B3MHX0 A0A1B6M0F8 B4MYJ1 A0A0K8TMZ0 A0A034VVZ2 A0A1B6HJX2 A0A0A1X224 W8ARW0 A0A1B6K1W3 A0A1B6FRP5 A0A0P9AF78 A0A1B6L3M9 A0A1B6HEU3 A0A1B0CQE2 B3NLL3 W8AVR1 A0A1W4VCB4 A0A0K8V9V0 A0A034W048 A0A0J9RFF1 B4P522 A0A182FMR7 E2QCH2 Q9U6B8 A0A023F5K7 N6V920 A0A1B6FIK9 A0A0V0GBU1 A0A069DXE3 A0A1I8PVS4 B4J9K5 A0A224XSH1 A0A3B0J5I1 T1P9I5 D3TQ77 A0A0P4VLK6 U5EU49 A0A0Q5VSX6 Q9U6B8-3 B4HN91 B4QBS3 A0A1A9XNG8 A0A1B0B0A2 A0A1W4V019 A0A1A9WHE0 A0A1L8DFL5 A0A0R1DS50 T1HN87
A0A2H1X1P9 A0A194RNY3 A0A194QHP9 A0A212FPV1 A0A0L7LFC4 A0A1E1W0N2 A0A2J7PD50 A0A1B6BWH1 A0A067RKQ5 A0A2A3E349 V9IMF7 A0A1B6DQT6 A0A1B6DJR3 A0A0C9S228 E0VCV9 A0A0Q9WFR7 E1ZXA4 B4KQU4 K7IRN4 A0A0N0BC83 A0A154PPF8 A0A0L7R8M8 A0A1B6DTE8 A0A195FT39 A0A195BD97 A0A195D4X5 A0A195DI95 A0A3L8E2D9 A0A026WLE9 B4LL00 A0A088A6F8 A0A158NQQ6 E2BPB3 A0A0Q9XLG5 F4WBV4 A0A151XB59 A0A0M4EFW8 A0A1J1IU05 E9IG63 A0A232F4J2 A0A023ENZ4 A0A182GIF4 A0A023EP20 Q1DGX6 A0A336MRH2 Q17KQ6 B3MHX0 A0A1B6M0F8 B4MYJ1 A0A0K8TMZ0 A0A034VVZ2 A0A1B6HJX2 A0A0A1X224 W8ARW0 A0A1B6K1W3 A0A1B6FRP5 A0A0P9AF78 A0A1B6L3M9 A0A1B6HEU3 A0A1B0CQE2 B3NLL3 W8AVR1 A0A1W4VCB4 A0A0K8V9V0 A0A034W048 A0A0J9RFF1 B4P522 A0A182FMR7 E2QCH2 Q9U6B8 A0A023F5K7 N6V920 A0A1B6FIK9 A0A0V0GBU1 A0A069DXE3 A0A1I8PVS4 B4J9K5 A0A224XSH1 A0A3B0J5I1 T1P9I5 D3TQ77 A0A0P4VLK6 U5EU49 A0A0Q5VSX6 Q9U6B8-3 B4HN91 B4QBS3 A0A1A9XNG8 A0A1B0B0A2 A0A1W4V019 A0A1A9WHE0 A0A1L8DFL5 A0A0R1DS50 T1HN87
Pubmed
19740753
19121390
26354079
22118469
26227816
24845553
+ More
20566863 17994087 20798317 20075255 30249741 24508170 21347285 21719571 21282665 28648823 24945155 26483478 17510324 18057021 26369729 25348373 25830018 24495485 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16751269 12537569 16582901 16645049 25474469 15632085 23185243 26334808 20353571 27129103
20566863 17994087 20798317 20075255 30249741 24508170 21347285 21719571 21282665 28648823 24945155 26483478 17510324 18057021 26369729 25348373 25830018 24495485 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16751269 12537569 16582901 16645049 25474469 15632085 23185243 26334808 20353571 27129103
EMBL
AB425231
BAG69139.1
BABH01008506
AB425232
BAG69140.1
NWSH01002969
+ More
PCG67178.1 PCG67179.1 KY174952 ATY75505.1 PCG67177.1 ODYU01012769 SOQ59241.1 KQ459984 KPJ19030.1 KQ458793 KPJ05068.1 AGBW02000411 OWR55776.1 JTDY01001350 KOB74110.1 GDQN01010509 JAT80545.1 NEVH01026394 PNF14257.1 GEDC01031667 JAS05631.1 KK852620 KDR20084.1 KZ288405 PBC26147.1 JR051951 AEY61594.1 GEDC01009261 JAS28037.1 GEDC01011362 JAS25936.1 GBYB01014931 JAG84698.1 DS235065 EEB11215.1 CH940648 KRF80244.1 GL435030 EFN74194.1 CH933808 EDW09293.2 KQ435922 KOX68562.1 KQ434998 KZC13324.1 KQ414632 KOC67183.1 GEDC01008357 JAS28941.1 KQ981280 KYN43467.1 KQ976511 KYM82533.1 KQ976885 KYN07479.1 KQ980824 KYN12557.1 QOIP01000001 RLU26864.1 KK107154 EZA56877.1 EDW61807.2 ADTU01023464 ADTU01023465 ADTU01023466 GL449574 EFN82468.1 KRG04579.1 GL888066 EGI68382.1 KQ982335 KYQ57584.1 CP012524 ALC42326.1 CVRI01000059 CRL03671.1 GL762910 EFZ20471.1 NNAY01001042 OXU25383.1 GAPW01002917 JAC10681.1 JXUM01066229 JXUM01066230 JXUM01066231 KQ562387 KXJ76013.1 GAPW01002893 JAC10705.1 CH900090 EAT32406.1 UFQS01001137 UFQT01001137 UFQT01001985 SSX09181.1 SSX32305.1 CH477222 EAT47248.1 CH902619 EDV36957.2 KPU76538.1 GEBQ01010607 GEBQ01007271 GEBQ01007103 GEBQ01003179 JAT29370.1 JAT32706.1 JAT32874.1 JAT36798.1 CH963894 EDW77180.2 GDAI01002080 JAI15523.1 GAKP01011441 GAKP01011440 GAKP01011439 JAC47511.1 GECU01032754 JAS74952.1 GBXI01009366 JAD04926.1 GAMC01017743 JAB88812.1 GECU01002281 JAT05426.1 GECZ01016911 JAS52858.1 KPU76537.1 GEBQ01021721 GEBQ01002336 JAT18256.1 JAT37641.1 GECU01034544 JAS73162.1 AJWK01023464 CH954179 EDV54988.1 KQS62052.1 GAMC01017742 JAB88813.1 GDHF01016701 GDHF01008376 JAI35613.1 JAI43938.1 GAKP01011442 JAC47510.1 CM002911 KMY94695.1 KMY94696.1 KMY94697.1 KMY94698.1 CM000158 EDW91723.1 KRK00046.1 KRK00047.1 AE013599 AAM68473.1 DQ503470 AF188634 AY071273 BT001876 BT003591 GBBI01002257 JAC16455.1 CM000071 ENO01906.2 KRT03044.1 GECZ01019723 JAS50046.1 GECL01000884 JAP05240.1 GBGD01002870 JAC86019.1 CH916367 EDW02512.1 GFTR01003658 JAW12768.1 OUUW01000001 SPP74922.1 KA645436 KA649240 AFP60065.1 EZ423579 ADD19855.1 GDKW01003274 JAI53321.1 GANO01002483 JAB57388.1 KQS62053.1 CH480816 EDW48373.1 CM000362 EDX07583.1 KMY94694.1 JXJN01006619 JXJN01006620 GFDF01008917 JAV05167.1 KRK00048.1 ACPB03020356
PCG67178.1 PCG67179.1 KY174952 ATY75505.1 PCG67177.1 ODYU01012769 SOQ59241.1 KQ459984 KPJ19030.1 KQ458793 KPJ05068.1 AGBW02000411 OWR55776.1 JTDY01001350 KOB74110.1 GDQN01010509 JAT80545.1 NEVH01026394 PNF14257.1 GEDC01031667 JAS05631.1 KK852620 KDR20084.1 KZ288405 PBC26147.1 JR051951 AEY61594.1 GEDC01009261 JAS28037.1 GEDC01011362 JAS25936.1 GBYB01014931 JAG84698.1 DS235065 EEB11215.1 CH940648 KRF80244.1 GL435030 EFN74194.1 CH933808 EDW09293.2 KQ435922 KOX68562.1 KQ434998 KZC13324.1 KQ414632 KOC67183.1 GEDC01008357 JAS28941.1 KQ981280 KYN43467.1 KQ976511 KYM82533.1 KQ976885 KYN07479.1 KQ980824 KYN12557.1 QOIP01000001 RLU26864.1 KK107154 EZA56877.1 EDW61807.2 ADTU01023464 ADTU01023465 ADTU01023466 GL449574 EFN82468.1 KRG04579.1 GL888066 EGI68382.1 KQ982335 KYQ57584.1 CP012524 ALC42326.1 CVRI01000059 CRL03671.1 GL762910 EFZ20471.1 NNAY01001042 OXU25383.1 GAPW01002917 JAC10681.1 JXUM01066229 JXUM01066230 JXUM01066231 KQ562387 KXJ76013.1 GAPW01002893 JAC10705.1 CH900090 EAT32406.1 UFQS01001137 UFQT01001137 UFQT01001985 SSX09181.1 SSX32305.1 CH477222 EAT47248.1 CH902619 EDV36957.2 KPU76538.1 GEBQ01010607 GEBQ01007271 GEBQ01007103 GEBQ01003179 JAT29370.1 JAT32706.1 JAT32874.1 JAT36798.1 CH963894 EDW77180.2 GDAI01002080 JAI15523.1 GAKP01011441 GAKP01011440 GAKP01011439 JAC47511.1 GECU01032754 JAS74952.1 GBXI01009366 JAD04926.1 GAMC01017743 JAB88812.1 GECU01002281 JAT05426.1 GECZ01016911 JAS52858.1 KPU76537.1 GEBQ01021721 GEBQ01002336 JAT18256.1 JAT37641.1 GECU01034544 JAS73162.1 AJWK01023464 CH954179 EDV54988.1 KQS62052.1 GAMC01017742 JAB88813.1 GDHF01016701 GDHF01008376 JAI35613.1 JAI43938.1 GAKP01011442 JAC47510.1 CM002911 KMY94695.1 KMY94696.1 KMY94697.1 KMY94698.1 CM000158 EDW91723.1 KRK00046.1 KRK00047.1 AE013599 AAM68473.1 DQ503470 AF188634 AY071273 BT001876 BT003591 GBBI01002257 JAC16455.1 CM000071 ENO01906.2 KRT03044.1 GECZ01019723 JAS50046.1 GECL01000884 JAP05240.1 GBGD01002870 JAC86019.1 CH916367 EDW02512.1 GFTR01003658 JAW12768.1 OUUW01000001 SPP74922.1 KA645436 KA649240 AFP60065.1 EZ423579 ADD19855.1 GDKW01003274 JAI53321.1 GANO01002483 JAB57388.1 KQS62053.1 CH480816 EDW48373.1 CM000362 EDX07583.1 KMY94694.1 JXJN01006619 JXJN01006620 GFDF01008917 JAV05167.1 KRK00048.1 ACPB03020356
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000235965 UP000027135 UP000242457 UP000009046 UP000008792 UP000000311 UP000009192 UP000002358 UP000053105 UP000076502 UP000053825 UP000078541 UP000078540 UP000078542 UP000078492 UP000279307 UP000053097 UP000005203 UP000005205 UP000008237 UP000007755 UP000075809 UP000092553 UP000183832 UP000215335 UP000069940 UP000249989 UP000008820 UP000007801 UP000007798 UP000092461 UP000008711 UP000192221 UP000002282 UP000069272 UP000000803 UP000001819 UP000095300 UP000001070 UP000268350 UP000001292 UP000000304 UP000092443 UP000092460 UP000091820 UP000015103
UP000235965 UP000027135 UP000242457 UP000009046 UP000008792 UP000000311 UP000009192 UP000002358 UP000053105 UP000076502 UP000053825 UP000078541 UP000078540 UP000078542 UP000078492 UP000279307 UP000053097 UP000005203 UP000005205 UP000008237 UP000007755 UP000075809 UP000092553 UP000183832 UP000215335 UP000069940 UP000249989 UP000008820 UP000007801 UP000007798 UP000092461 UP000008711 UP000192221 UP000002282 UP000069272 UP000000803 UP000001819 UP000095300 UP000001070 UP000268350 UP000001292 UP000000304 UP000092443 UP000092460 UP000091820 UP000015103
Pfam
PF07856 Orai-1
Gene 3D
ProteinModelPortal
B5BRC4
B5BRC5
A0A2A4J677
A0A2A4J5M0
A0A2H4THI5
A0A2A4J4V7
+ More
A0A2H1X1P9 A0A194RNY3 A0A194QHP9 A0A212FPV1 A0A0L7LFC4 A0A1E1W0N2 A0A2J7PD50 A0A1B6BWH1 A0A067RKQ5 A0A2A3E349 V9IMF7 A0A1B6DQT6 A0A1B6DJR3 A0A0C9S228 E0VCV9 A0A0Q9WFR7 E1ZXA4 B4KQU4 K7IRN4 A0A0N0BC83 A0A154PPF8 A0A0L7R8M8 A0A1B6DTE8 A0A195FT39 A0A195BD97 A0A195D4X5 A0A195DI95 A0A3L8E2D9 A0A026WLE9 B4LL00 A0A088A6F8 A0A158NQQ6 E2BPB3 A0A0Q9XLG5 F4WBV4 A0A151XB59 A0A0M4EFW8 A0A1J1IU05 E9IG63 A0A232F4J2 A0A023ENZ4 A0A182GIF4 A0A023EP20 Q1DGX6 A0A336MRH2 Q17KQ6 B3MHX0 A0A1B6M0F8 B4MYJ1 A0A0K8TMZ0 A0A034VVZ2 A0A1B6HJX2 A0A0A1X224 W8ARW0 A0A1B6K1W3 A0A1B6FRP5 A0A0P9AF78 A0A1B6L3M9 A0A1B6HEU3 A0A1B0CQE2 B3NLL3 W8AVR1 A0A1W4VCB4 A0A0K8V9V0 A0A034W048 A0A0J9RFF1 B4P522 A0A182FMR7 E2QCH2 Q9U6B8 A0A023F5K7 N6V920 A0A1B6FIK9 A0A0V0GBU1 A0A069DXE3 A0A1I8PVS4 B4J9K5 A0A224XSH1 A0A3B0J5I1 T1P9I5 D3TQ77 A0A0P4VLK6 U5EU49 A0A0Q5VSX6 Q9U6B8-3 B4HN91 B4QBS3 A0A1A9XNG8 A0A1B0B0A2 A0A1W4V019 A0A1A9WHE0 A0A1L8DFL5 A0A0R1DS50 T1HN87
A0A2H1X1P9 A0A194RNY3 A0A194QHP9 A0A212FPV1 A0A0L7LFC4 A0A1E1W0N2 A0A2J7PD50 A0A1B6BWH1 A0A067RKQ5 A0A2A3E349 V9IMF7 A0A1B6DQT6 A0A1B6DJR3 A0A0C9S228 E0VCV9 A0A0Q9WFR7 E1ZXA4 B4KQU4 K7IRN4 A0A0N0BC83 A0A154PPF8 A0A0L7R8M8 A0A1B6DTE8 A0A195FT39 A0A195BD97 A0A195D4X5 A0A195DI95 A0A3L8E2D9 A0A026WLE9 B4LL00 A0A088A6F8 A0A158NQQ6 E2BPB3 A0A0Q9XLG5 F4WBV4 A0A151XB59 A0A0M4EFW8 A0A1J1IU05 E9IG63 A0A232F4J2 A0A023ENZ4 A0A182GIF4 A0A023EP20 Q1DGX6 A0A336MRH2 Q17KQ6 B3MHX0 A0A1B6M0F8 B4MYJ1 A0A0K8TMZ0 A0A034VVZ2 A0A1B6HJX2 A0A0A1X224 W8ARW0 A0A1B6K1W3 A0A1B6FRP5 A0A0P9AF78 A0A1B6L3M9 A0A1B6HEU3 A0A1B0CQE2 B3NLL3 W8AVR1 A0A1W4VCB4 A0A0K8V9V0 A0A034W048 A0A0J9RFF1 B4P522 A0A182FMR7 E2QCH2 Q9U6B8 A0A023F5K7 N6V920 A0A1B6FIK9 A0A0V0GBU1 A0A069DXE3 A0A1I8PVS4 B4J9K5 A0A224XSH1 A0A3B0J5I1 T1P9I5 D3TQ77 A0A0P4VLK6 U5EU49 A0A0Q5VSX6 Q9U6B8-3 B4HN91 B4QBS3 A0A1A9XNG8 A0A1B0B0A2 A0A1W4V019 A0A1A9WHE0 A0A1L8DFL5 A0A0R1DS50 T1HN87
PDB
6BBG
E-value=5.01593e-64,
Score=617
Ontologies
GO
PANTHER
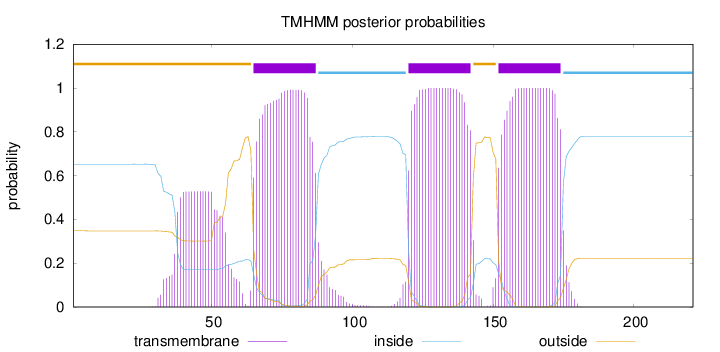
Topology
Subcellular location
Cell membrane
Length:
221
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
77.94872
Exp number, first 60 AAs:
10.42785
Total prob of N-in:
0.65225
POSSIBLE N-term signal
sequence
outside
1 - 64
TMhelix
65 - 87
inside
88 - 119
TMhelix
120 - 142
outside
143 - 151
TMhelix
152 - 174
inside
175 - 221
Population Genetic Test Statistics
Pi
200.451751
Theta
162.881095
Tajima's D
0.009993
CLR
345.342034
CSRT
0.372781360931953
Interpretation
Uncertain
