Gene
KWMTBOMO05509 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006722
Annotation
PREDICTED:_28S_ribosomal_protein_S22?_mitochondrial_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.291
Sequence
CDS
ATGTCTTTACTGTTACAAAAAATTGCAACGAAGCATGTTAAAACATTAATAATAAAATATGAGCCGTGCGTTCTAGTTATATCCAATCGAAAATTGAACATTGTACCATCATTTTACGATGGCGAACAAGACCCGGCTCCAAAGTTTTTTACACCAAATGTACAGAATCTATTAAAGAGATTGACTCGGCCAGATTTTACTAAAGTTTTCCGCAAGAGAAACAATCAAGGTCTACATGCCCTTAAAACTCCCAAATATAAGTTTCTCACGAAAGAAGAGCTTGAATTGGAAATTGCTAAAGCTAATGAAAAAGCCGATGTGCTTCTTCAAATACCTCCTGTCCTCAAAATTCACAAACCCATTGATGAGGTTTTATCAAGAGACCCAGCTTTGATAGGATATGACACATCAAAATATTTGTTTACCGACATCACATTTGGTATTGCCAATGAAGAGAGGATTATTATTGAAAGAGACACTGATGGAACTCTGCGCAGTTGTGATCATGATATCAGAAAGAAATTAAATCAGGTGTACTTTCCAATAACCGGACGCAAAATAAGAGAACCACACATGTTTTCTGATGAGGAAAAATTTCACAATCTGCTGGACAGGGGAGAGTACGAGTTTGTTCTGGATCGAGCCTGCATACAATATGAACCGGACGAACCAATATTCCAAAAGATTACCAGTATTACATACCAGCATGTGGACAATAATAAATGTTACAACACATTAAGATCAACAAGACACTTTGGTTCAATGACTTTTTACCTGACATGGCATCAGAGTATTGATAATCTGATGCTCGAGATCATACAAAGCGGTTTCATCAGAGAGGCGGTTTTGCTTGCTGCGCTACGGCACGATGTTCACAACGACGTGAGGGAAGGTACCGCGGCACAGTCTTTAGCTCAAGAAATTTTTACCACACCTCTGCTGCTATCTAAACTAGAACGTCTCACTGAGGAGGACATTCAGCTTGACAGTAAATGTATTCAGTGCATAGAGAAATATATAGTGAGCAACTCGACGATAAAGAGTCAACATGAATTAGCGCTGCAAGGCTTCAAAGAAGATTTCCAGCACCTCTCCGAGCTTAGAAAGGGTTTAGAAAAAGCTCATGGAAACGTTTAA
Protein
MSLLLQKIATKHVKTLIIKYEPCVLVISNRKLNIVPSFYDGEQDPAPKFFTPNVQNLLKRLTRPDFTKVFRKRNNQGLHALKTPKYKFLTKEELELEIAKANEKADVLLQIPPVLKIHKPIDEVLSRDPALIGYDTSKYLFTDITFGIANEERIIIERDTDGTLRSCDHDIRKKLNQVYFPITGRKIREPHMFSDEEKFHNLLDRGEYEFVLDRACIQYEPDEPIFQKITSITYQHVDNNKCYNTLRSTRHFGSMTFYLTWHQSIDNLMLEIIQSGFIREAVLLAALRHDVHNDVREGTAAQSLAQEIFTTPLLLSKLERLTEEDIQLDSKCIQCIEKYIVSNSTIKSQHELALQGFKEDFQHLSELRKGLEKAHGNV
Summary
Uniprot
H9JB27
A0A2A4J520
A0A2H1WLN3
A0A1E1WQJ0
A0A3S2L2W8
I4DPQ6
+ More
A0A212FPT7 A0A194QNK5 D6WJ69 A0A1W4WH68 A0A0P4W475 A0A067QRU5 A0A0C9RWS6 A0A2J7QVH5 N6UCS6 U4U4G9 A0A182TJ66 A0A1Y1MBC2 A0NBG6 A0A182X3M8 A0A1Q3F7M1 Q17HE9 A0A182I3E1 A0A182V0Q8 A0A182IN91 A0A182JQP5 A0A182XYZ2 B0WIA1 A0A182KRG2 A0A182TBW8 A0A084WNE7 A0A182PQC0 A0A182G350 A0A023EQM8 A0A182MBN3 A0A182NHS4 A0A182WJ55 A0A0T6AW02 A0A182R4E4 A0A182QXV7 A0A3L8D8E2 A0A1I8MB78 A0A1I8PB53 A0A026WIV3 T1PAJ3 A0A1B0DNZ0 A0A2P8YCG4 B3LX12 A0A2M4AQL0 A0A0J7N9U6 A0A2M4BRC8 A0A2M4AQT9 A0A182FF73 B4KAQ8 A0A2M4AQ95 A0A336MAH8 A0A2M3ZE03 B4NAT3 U5EW17 B4GNU9 Q29BE9 A0A2M4BRM0 A0A1B0BUZ8 A0A232F7I2 A0A0K8TSG5 A0A1A9ULY0 A0A1A9Y2J4 A0A3B0JK81 A0A0M4EK51 A0A1L8E080 A0A0L0CJK2 W5JGZ4 K7J694 A0A1A9Z7A2 A0A182TWZ9 A0A1W4VMF2 B4M602 A0A1B0FP52 A0A087ZSI4 A0A1A9WBZ8 A0A195BNA5 B4QY50 A0A158NRQ1 B4IGX2 Q9VAY9 A0A0A1X8C8 A0A2A3EF30 B3P5W4 A0A034WFA6 A0A1B6G308 A0A1B6IFX5 A0A2P2HYE8 A0A1B6HIQ6 W8BD94
A0A212FPT7 A0A194QNK5 D6WJ69 A0A1W4WH68 A0A0P4W475 A0A067QRU5 A0A0C9RWS6 A0A2J7QVH5 N6UCS6 U4U4G9 A0A182TJ66 A0A1Y1MBC2 A0NBG6 A0A182X3M8 A0A1Q3F7M1 Q17HE9 A0A182I3E1 A0A182V0Q8 A0A182IN91 A0A182JQP5 A0A182XYZ2 B0WIA1 A0A182KRG2 A0A182TBW8 A0A084WNE7 A0A182PQC0 A0A182G350 A0A023EQM8 A0A182MBN3 A0A182NHS4 A0A182WJ55 A0A0T6AW02 A0A182R4E4 A0A182QXV7 A0A3L8D8E2 A0A1I8MB78 A0A1I8PB53 A0A026WIV3 T1PAJ3 A0A1B0DNZ0 A0A2P8YCG4 B3LX12 A0A2M4AQL0 A0A0J7N9U6 A0A2M4BRC8 A0A2M4AQT9 A0A182FF73 B4KAQ8 A0A2M4AQ95 A0A336MAH8 A0A2M3ZE03 B4NAT3 U5EW17 B4GNU9 Q29BE9 A0A2M4BRM0 A0A1B0BUZ8 A0A232F7I2 A0A0K8TSG5 A0A1A9ULY0 A0A1A9Y2J4 A0A3B0JK81 A0A0M4EK51 A0A1L8E080 A0A0L0CJK2 W5JGZ4 K7J694 A0A1A9Z7A2 A0A182TWZ9 A0A1W4VMF2 B4M602 A0A1B0FP52 A0A087ZSI4 A0A1A9WBZ8 A0A195BNA5 B4QY50 A0A158NRQ1 B4IGX2 Q9VAY9 A0A0A1X8C8 A0A2A3EF30 B3P5W4 A0A034WFA6 A0A1B6G308 A0A1B6IFX5 A0A2P2HYE8 A0A1B6HIQ6 W8BD94
Pubmed
19121390
22651552
22118469
26354079
18362917
19820115
+ More
24845553 23537049 28004739 12364791 14747013 17210077 17510324 25244985 20966253 24438588 26483478 24945155 30249741 25315136 24508170 29403074 17994087 15632085 28648823 26369729 26108605 20920257 23761445 20075255 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 24495485
24845553 23537049 28004739 12364791 14747013 17210077 17510324 25244985 20966253 24438588 26483478 24945155 30249741 25315136 24508170 29403074 17994087 15632085 28648823 26369729 26108605 20920257 23761445 20075255 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 24495485
EMBL
BABH01008506
NWSH01002969
PCG67175.1
ODYU01009490
SOQ53969.1
GDQN01001963
+ More
JAT89091.1 RSAL01000201 RVE44437.1 AK403722 BAM19896.1 AGBW02000411 OWR55778.1 KQ458793 KPJ05066.1 KQ971343 EFA04432.1 GDRN01093318 GDRN01093317 GDRN01093316 JAI59907.1 KK853097 KDR11447.1 GBYB01012280 GBYB01012281 JAG82047.1 JAG82048.1 NEVH01010475 PNF32587.1 APGK01034106 KB740904 ENN78436.1 KB632006 ERL87947.1 GEZM01036125 JAV82953.1 AAAB01008807 EAU77614.2 GFDL01011489 JAV23556.1 CH477250 EAT46057.1 APCN01000207 DS231945 EDS28327.1 ATLV01024593 KE525352 KFB51741.1 JXUM01140965 KQ569159 KXJ68747.1 GAPW01002203 JAC11395.1 AXCM01000285 LJIG01022672 KRT79324.1 AXCN02000377 QOIP01000011 RLU16564.1 KK107183 EZA55913.1 KA645175 AFP59804.1 AJVK01007866 PYGN01000705 PSN41940.1 CH902617 EDV43851.1 GGFK01009756 MBW43077.1 LBMM01007760 KMQ89445.1 GGFJ01006431 MBW55572.1 GGFK01009647 MBW42968.1 CH933806 EDW16795.1 GGFK01009618 MBW42939.1 UFQS01000305 UFQT01000305 SSX02674.1 SSX23048.1 GGFM01006033 MBW26784.1 CH964232 EDW80897.1 GANO01003161 JAB56710.1 CH479186 EDW38832.1 CM000070 EAL27049.1 GGFJ01006430 MBW55571.1 JXJN01020989 NNAY01000798 OXU26450.1 GDAI01000525 JAI17078.1 OUUW01000005 SPP80732.1 CP012526 ALC47484.1 GFDF01001983 JAV12101.1 JRES01000310 KNC32425.1 ADMH02001422 ETN62593.1 CH940652 EDW59078.1 CCAG010023115 KQ976439 KYM86681.1 CM000364 EDX14698.1 ADTU01024219 CH480837 EDW49090.1 AE014297 BT021953 AAF56757.1 AAX51658.1 GBXI01007297 JAD06995.1 KZ288280 PBC29621.1 CH954182 EDV53364.1 GAKP01006137 JAC52815.1 GECZ01012944 JAS56825.1 GECU01021885 JAS85821.1 IACF01001082 LAB66804.1 GECU01033164 JAS74542.1 GAMC01007285 JAB99270.1
JAT89091.1 RSAL01000201 RVE44437.1 AK403722 BAM19896.1 AGBW02000411 OWR55778.1 KQ458793 KPJ05066.1 KQ971343 EFA04432.1 GDRN01093318 GDRN01093317 GDRN01093316 JAI59907.1 KK853097 KDR11447.1 GBYB01012280 GBYB01012281 JAG82047.1 JAG82048.1 NEVH01010475 PNF32587.1 APGK01034106 KB740904 ENN78436.1 KB632006 ERL87947.1 GEZM01036125 JAV82953.1 AAAB01008807 EAU77614.2 GFDL01011489 JAV23556.1 CH477250 EAT46057.1 APCN01000207 DS231945 EDS28327.1 ATLV01024593 KE525352 KFB51741.1 JXUM01140965 KQ569159 KXJ68747.1 GAPW01002203 JAC11395.1 AXCM01000285 LJIG01022672 KRT79324.1 AXCN02000377 QOIP01000011 RLU16564.1 KK107183 EZA55913.1 KA645175 AFP59804.1 AJVK01007866 PYGN01000705 PSN41940.1 CH902617 EDV43851.1 GGFK01009756 MBW43077.1 LBMM01007760 KMQ89445.1 GGFJ01006431 MBW55572.1 GGFK01009647 MBW42968.1 CH933806 EDW16795.1 GGFK01009618 MBW42939.1 UFQS01000305 UFQT01000305 SSX02674.1 SSX23048.1 GGFM01006033 MBW26784.1 CH964232 EDW80897.1 GANO01003161 JAB56710.1 CH479186 EDW38832.1 CM000070 EAL27049.1 GGFJ01006430 MBW55571.1 JXJN01020989 NNAY01000798 OXU26450.1 GDAI01000525 JAI17078.1 OUUW01000005 SPP80732.1 CP012526 ALC47484.1 GFDF01001983 JAV12101.1 JRES01000310 KNC32425.1 ADMH02001422 ETN62593.1 CH940652 EDW59078.1 CCAG010023115 KQ976439 KYM86681.1 CM000364 EDX14698.1 ADTU01024219 CH480837 EDW49090.1 AE014297 BT021953 AAF56757.1 AAX51658.1 GBXI01007297 JAD06995.1 KZ288280 PBC29621.1 CH954182 EDV53364.1 GAKP01006137 JAC52815.1 GECZ01012944 JAS56825.1 GECU01021885 JAS85821.1 IACF01001082 LAB66804.1 GECU01033164 JAS74542.1 GAMC01007285 JAB99270.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000007266
+ More
UP000192223 UP000027135 UP000235965 UP000019118 UP000030742 UP000075902 UP000007062 UP000076407 UP000008820 UP000075840 UP000075903 UP000075880 UP000075881 UP000076408 UP000002320 UP000075882 UP000075901 UP000030765 UP000075885 UP000069940 UP000249989 UP000075883 UP000075884 UP000075920 UP000075900 UP000075886 UP000279307 UP000095301 UP000095300 UP000053097 UP000092462 UP000245037 UP000007801 UP000036403 UP000069272 UP000009192 UP000007798 UP000008744 UP000001819 UP000092460 UP000215335 UP000078200 UP000092443 UP000268350 UP000092553 UP000037069 UP000000673 UP000002358 UP000092445 UP000192221 UP000008792 UP000092444 UP000005203 UP000091820 UP000078540 UP000000304 UP000005205 UP000001292 UP000000803 UP000242457 UP000008711
UP000192223 UP000027135 UP000235965 UP000019118 UP000030742 UP000075902 UP000007062 UP000076407 UP000008820 UP000075840 UP000075903 UP000075880 UP000075881 UP000076408 UP000002320 UP000075882 UP000075901 UP000030765 UP000075885 UP000069940 UP000249989 UP000075883 UP000075884 UP000075920 UP000075900 UP000075886 UP000279307 UP000095301 UP000095300 UP000053097 UP000092462 UP000245037 UP000007801 UP000036403 UP000069272 UP000009192 UP000007798 UP000008744 UP000001819 UP000092460 UP000215335 UP000078200 UP000092443 UP000268350 UP000092553 UP000037069 UP000000673 UP000002358 UP000092445 UP000192221 UP000008792 UP000092444 UP000005203 UP000091820 UP000078540 UP000000304 UP000005205 UP000001292 UP000000803 UP000242457 UP000008711
Pfam
PF10245 MRP-S22
Interpro
IPR019374
Ribosomal_S22_mit
ProteinModelPortal
H9JB27
A0A2A4J520
A0A2H1WLN3
A0A1E1WQJ0
A0A3S2L2W8
I4DPQ6
+ More
A0A212FPT7 A0A194QNK5 D6WJ69 A0A1W4WH68 A0A0P4W475 A0A067QRU5 A0A0C9RWS6 A0A2J7QVH5 N6UCS6 U4U4G9 A0A182TJ66 A0A1Y1MBC2 A0NBG6 A0A182X3M8 A0A1Q3F7M1 Q17HE9 A0A182I3E1 A0A182V0Q8 A0A182IN91 A0A182JQP5 A0A182XYZ2 B0WIA1 A0A182KRG2 A0A182TBW8 A0A084WNE7 A0A182PQC0 A0A182G350 A0A023EQM8 A0A182MBN3 A0A182NHS4 A0A182WJ55 A0A0T6AW02 A0A182R4E4 A0A182QXV7 A0A3L8D8E2 A0A1I8MB78 A0A1I8PB53 A0A026WIV3 T1PAJ3 A0A1B0DNZ0 A0A2P8YCG4 B3LX12 A0A2M4AQL0 A0A0J7N9U6 A0A2M4BRC8 A0A2M4AQT9 A0A182FF73 B4KAQ8 A0A2M4AQ95 A0A336MAH8 A0A2M3ZE03 B4NAT3 U5EW17 B4GNU9 Q29BE9 A0A2M4BRM0 A0A1B0BUZ8 A0A232F7I2 A0A0K8TSG5 A0A1A9ULY0 A0A1A9Y2J4 A0A3B0JK81 A0A0M4EK51 A0A1L8E080 A0A0L0CJK2 W5JGZ4 K7J694 A0A1A9Z7A2 A0A182TWZ9 A0A1W4VMF2 B4M602 A0A1B0FP52 A0A087ZSI4 A0A1A9WBZ8 A0A195BNA5 B4QY50 A0A158NRQ1 B4IGX2 Q9VAY9 A0A0A1X8C8 A0A2A3EF30 B3P5W4 A0A034WFA6 A0A1B6G308 A0A1B6IFX5 A0A2P2HYE8 A0A1B6HIQ6 W8BD94
A0A212FPT7 A0A194QNK5 D6WJ69 A0A1W4WH68 A0A0P4W475 A0A067QRU5 A0A0C9RWS6 A0A2J7QVH5 N6UCS6 U4U4G9 A0A182TJ66 A0A1Y1MBC2 A0NBG6 A0A182X3M8 A0A1Q3F7M1 Q17HE9 A0A182I3E1 A0A182V0Q8 A0A182IN91 A0A182JQP5 A0A182XYZ2 B0WIA1 A0A182KRG2 A0A182TBW8 A0A084WNE7 A0A182PQC0 A0A182G350 A0A023EQM8 A0A182MBN3 A0A182NHS4 A0A182WJ55 A0A0T6AW02 A0A182R4E4 A0A182QXV7 A0A3L8D8E2 A0A1I8MB78 A0A1I8PB53 A0A026WIV3 T1PAJ3 A0A1B0DNZ0 A0A2P8YCG4 B3LX12 A0A2M4AQL0 A0A0J7N9U6 A0A2M4BRC8 A0A2M4AQT9 A0A182FF73 B4KAQ8 A0A2M4AQ95 A0A336MAH8 A0A2M3ZE03 B4NAT3 U5EW17 B4GNU9 Q29BE9 A0A2M4BRM0 A0A1B0BUZ8 A0A232F7I2 A0A0K8TSG5 A0A1A9ULY0 A0A1A9Y2J4 A0A3B0JK81 A0A0M4EK51 A0A1L8E080 A0A0L0CJK2 W5JGZ4 K7J694 A0A1A9Z7A2 A0A182TWZ9 A0A1W4VMF2 B4M602 A0A1B0FP52 A0A087ZSI4 A0A1A9WBZ8 A0A195BNA5 B4QY50 A0A158NRQ1 B4IGX2 Q9VAY9 A0A0A1X8C8 A0A2A3EF30 B3P5W4 A0A034WFA6 A0A1B6G308 A0A1B6IFX5 A0A2P2HYE8 A0A1B6HIQ6 W8BD94
PDB
6GAZ
E-value=3.17103e-44,
Score=449
Ontologies
PANTHER
Topology
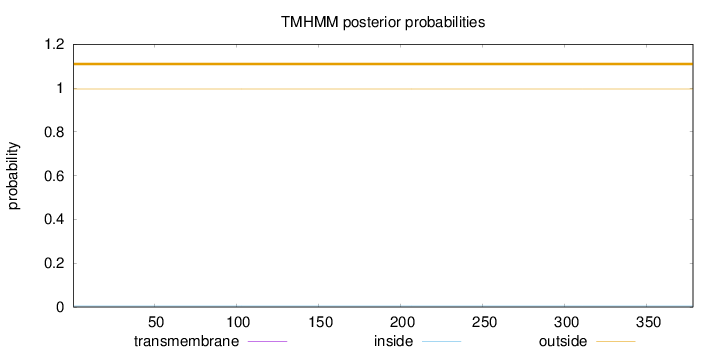
Length:
378
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00231
Exp number, first 60 AAs:
0.00053
Total prob of N-in:
0.00484
outside
1 - 378
Population Genetic Test Statistics
Pi
181.285304
Theta
178.670895
Tajima's D
-0.264017
CLR
0.871572
CSRT
0.298085095745213
Interpretation
Uncertain
