Gene
KWMTBOMO05506
Pre Gene Modal
BGIBMGA006619
Annotation
dopamine_transporter_[Bombyx_mori]
Full name
Transporter
+ More
Sodium-dependent dopamine transporter
Sodium-dependent dopamine transporter
Alternative Name
Protein fumin
Location in the cell
PlasmaMembrane Reliability : 4.883
Sequence
CDS
ATGGCGCTGAAGACCCCGACGCCGGGTGTTGTCGGCGAACGCGAGACCTGGGGCAAGAAGGTCGACTTCCTGCTCTCGGTCATCGGGTTCGCCGTCGACCTCGCCAACGTGTGGCGCTTCCCCTACCTCTGCTATAAGAATGGAGGGGGTGCGTTTTTGGTGCCATACTGCATCATGCTGGTAGTGGGTGGAATTCCACTTTTCTACATGGAGTTGGCGCTGGGACAATTTCACAGGAAGGGTGCCATCACCTGCTGGGGCAGGCTTGTCCCGCTTTTCAAAGGAATCGGTTATGCTGTCGTCCTGATCGCGTTCTACGTGGACTTTTACTACAACGTGATAATCGCTTGGGCGCTGCGTTTTTTCTTCGCCTCTTTCACGACCATGCTACCTTGGACCAACTGCGACAACGAATGGAACACACCTGCCTGCCGACCGTTCGAAGCCATTTGGGACGTTAACCGTACACGTATACGCAATACCACGTCCGCAAGTTTGGGAATCGCTCCGACAACACCGTACACTTCTGCTGCTTCGGAATATTTCAACCGCGCGATTTTAGAATTGCAAGGCAGCGAGGGGCTCCACGACCTTGGTTCTGTAAAATGGGATATGGCGCTGTGTTTATTGGCCGTCTACGTCATCTGCTACTTCTCCTTGTGGAAGGGAATCAGTACTTCTGGAAAAGTTGTTTGGTTCACAGCACTCTTTCCGTACGCTGTTCTTTTGATACTCTTGGTTCGTGGTATCACCCTCCCTGGATCGGCGACCGGAATCCAATACTATCTGAGTCCGAACTTTGAAGCTATAACTCAACCTCAGGTGTGGGTGGACGCCGCTACTCAAGTCTTCTTTTCTCTTGGACCGGGCTTTGGAGTGCTCTTGGCTTATGCATCGTACAACAAGTACCACAACAACGTGTACAAGGACGCCATTTTGACCAGCGTCATAAATTCCGCAACGTCTTTCGTCGCTGGTTTCGTGATCTTCAGCGTGCTCGGCTACATGGCGCACGCTTCGGGCAGAGACGTTCAGGATGTCGCAACCGAAGGCCCGGGACTAGTTTTCGTGGTGTACCCGGCGGCTATAGCTACGATGCCAGGATCAACGTTTTGGGCGCTTATATTCTTCATGATGCTGCTTACACTTGGGCTTGATAGTTCTTTTGGGGGTTCCGAAGCGATAATCACAGCTCTCAGTGACGAGTTCCCTCCTATCGGACGTCATCGCGAATTGTTCGTCGCCTGCCTCTTTACGTTGTATTTCTTCGTGGGACTGGCCTCTTGCACGAAAGGCGGATTTTACTTCTTCCAGCTCTTGGATCGCTATGCTGCGGGCTATTCTATTCTCATCGCTGTTTTCTTTGAGGCCATAGCTGTCTCCTGGATTTATGGCACGGAAAGATTTTGTGAGGACATTCGCGACATGATCGGCTTCAGACCTGGCCTGTACTGGCGCGTTTGTTGGCGTTTCGCGGCACCATCTTTTTTGCTCTTCATCACGGCATACGGACTCTTAGACTACGAACCTCTGCAATACGAGAACTACATCTATCCAGGCTGGGCCAACGCACTTGGCTGGGCCATTGCTGGATCCAGCGTCATGTGCATACCGACAGTGGCTATCTATAAACTGATTACAACTAAAGGATCGTTTTTGGAGCGTCTTCGCGTTTTAACAACCCCTTATGCAGACAGCGAACGTAATGGAACCGTGCACAACGGTATGATAGTGTCCGAGAGCGGCGGCGTTCGGCTGACGTCAGCGGTGCAAACCCCCACAACTCCACAGCAACCGATCGGAGCAAATATTCCAGCAGCCTCGGCTCCAACTCTCGCTTCATCTCCAGCGCTGGTCTGA
Protein
MALKTPTPGVVGERETWGKKVDFLLSVIGFAVDLANVWRFPYLCYKNGGGAFLVPYCIMLVVGGIPLFYMELALGQFHRKGAITCWGRLVPLFKGIGYAVVLIAFYVDFYYNVIIAWALRFFFASFTTMLPWTNCDNEWNTPACRPFEAIWDVNRTRIRNTTSASLGIAPTTPYTSAASEYFNRAILELQGSEGLHDLGSVKWDMALCLLAVYVICYFSLWKGISTSGKVVWFTALFPYAVLLILLVRGITLPGSATGIQYYLSPNFEAITQPQVWVDAATQVFFSLGPGFGVLLAYASYNKYHNNVYKDAILTSVINSATSFVAGFVIFSVLGYMAHASGRDVQDVATEGPGLVFVVYPAAIATMPGSTFWALIFFMMLLTLGLDSSFGGSEAIITALSDEFPPIGRHRELFVACLFTLYFFVGLASCTKGGFYFFQLLDRYAAGYSILIAVFFEAIAVSWIYGTERFCEDIRDMIGFRPGLYWRVCWRFAAPSFLLFITAYGLLDYEPLQYENYIYPGWANALGWAIAGSSVMCIPTVAIYKLITTKGSFLERLRVLTTPYADSERNGTVHNGMIVSESGGVRLTSAVQTPTTPQQPIGANIPAASAPTLASSPALV
Summary
Description
Sodium-dependent dopamine transporter which terminates the action of dopamine by its high affinity sodium-dependent reuptake into presynaptic terminals (PubMed:11125028, PubMed:12606774, PubMed:24037379, PubMed:25970245). Also transports tyramine and norepinephrine, shows less efficient transport of octopamine and does not transport serotonin (PubMed:11125028, PubMed:12606774). Plays a role in the regulation of the rest/activity cycle (PubMed:16093388, PubMed:25232310).
Miscellaneous
10-fold less sensitive to cocaine than mammalian dopamine transporter SLC6A3.
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Keywords
3D-structure
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Membrane
Metal-binding
Neurotransmitter transport
Reference proteome
Sodium
Symport
Transmembrane
Transmembrane helix
Transport
Feature
chain Transporter
Uniprot
Q45VA3
A0A2A4IX52
Q86MC4
H9JAS4
A0A2H1WLW5
A0A212F0P2
+ More
Q45VA2 A0A194RSJ2 A0A0G3VG04 A0A3S2NTV6 A0A194QHP4 A0A0L7LLJ2 Q16TV9 A0A1I8MCE3 A0A0V0GBJ2 A0A023F4F4 I2FKF4 A0A069DV82 A0A224X9N0 A0A2J7RJL9 A0A0L0BLL6 A0A182IX90 A0A1I8PFW0 A0A195FSP1 E1ZXA1 A0A084WN18 A0A182QPG6 A0A195D3J0 B4KSI5 A0A158NQQ9 A0A151WND3 A0A182UWK2 Q7QJ94 F4WBV1 A0A195BE92 A0A1A9WR92 A0A195DI93 W8C6P2 B4P656 W8BA44 B3NPH4 B4LMF2 B4HSZ4 B4J7H0 B4QI04 A0A0B4KEX2 Q7K4Y6 A0A1B0FKI7 A0A154P078 A0A026WMI7 A0A3L8E4C5 A0A1W4UKV4 A0A1B6MBJ9 A0A2A3EG39 A0A3B0JJ43 A0A0M9A3V3 A0A087ZQ28 B4MPH7 A0A0K8VN76 B9A0V7 A0A0K8U8S5 Q291U3 B4GA93 A0A336MNN5 A0A0M5IXL3 A0A1J1ICN7 A0A0A1X506 A0A2H8TLX0 A0A2S2PRA9 E9HBL2 B0WUE7 T1I1H1 A0A0P5IDE2 A0A0P6BYM5 E9IG58 A0A182UCW0 A0A182FDF5 A0A182PRE6 A0A226E682 A0A182NI32 A0A182XYN8 A0A182MIW2 A0A182WJF0 A0A1B0B3H9 A0A310SJT6 A0A0P5LUT0 A0A182I335 A0A0P5RJ52 A0A182X3Y4 A0A182JRM0 W5JIV4 A0A087T7C5 A0A182R7A4 B4K361 T1PKA4 A0A0N8ACY8 A0A164NRL7 A0A1B6HM80 A0A182SRQ8 A0A2R5LK72
Q45VA2 A0A194RSJ2 A0A0G3VG04 A0A3S2NTV6 A0A194QHP4 A0A0L7LLJ2 Q16TV9 A0A1I8MCE3 A0A0V0GBJ2 A0A023F4F4 I2FKF4 A0A069DV82 A0A224X9N0 A0A2J7RJL9 A0A0L0BLL6 A0A182IX90 A0A1I8PFW0 A0A195FSP1 E1ZXA1 A0A084WN18 A0A182QPG6 A0A195D3J0 B4KSI5 A0A158NQQ9 A0A151WND3 A0A182UWK2 Q7QJ94 F4WBV1 A0A195BE92 A0A1A9WR92 A0A195DI93 W8C6P2 B4P656 W8BA44 B3NPH4 B4LMF2 B4HSZ4 B4J7H0 B4QI04 A0A0B4KEX2 Q7K4Y6 A0A1B0FKI7 A0A154P078 A0A026WMI7 A0A3L8E4C5 A0A1W4UKV4 A0A1B6MBJ9 A0A2A3EG39 A0A3B0JJ43 A0A0M9A3V3 A0A087ZQ28 B4MPH7 A0A0K8VN76 B9A0V7 A0A0K8U8S5 Q291U3 B4GA93 A0A336MNN5 A0A0M5IXL3 A0A1J1ICN7 A0A0A1X506 A0A2H8TLX0 A0A2S2PRA9 E9HBL2 B0WUE7 T1I1H1 A0A0P5IDE2 A0A0P6BYM5 E9IG58 A0A182UCW0 A0A182FDF5 A0A182PRE6 A0A226E682 A0A182NI32 A0A182XYN8 A0A182MIW2 A0A182WJF0 A0A1B0B3H9 A0A310SJT6 A0A0P5LUT0 A0A182I335 A0A0P5RJ52 A0A182X3Y4 A0A182JRM0 W5JIV4 A0A087T7C5 A0A182R7A4 B4K361 T1PKA4 A0A0N8ACY8 A0A164NRL7 A0A1B6HM80 A0A182SRQ8 A0A2R5LK72
Pubmed
16310975
19121390
22118469
26354079
26227816
17510324
+ More
25315136 25474469 26334808 26108605 20798317 24438588 17994087 21347285 12364791 21719571 24495485 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11125028 12606774 12537569 16093388 17131045 25232310 24037379 25970245 25961798 24508170 30249741 15632085 25830018 21292972 21282665 25244985 20920257 23761445
25315136 25474469 26334808 26108605 20798317 24438588 17994087 21347285 12364791 21719571 24495485 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11125028 12606774 12537569 16093388 17131045 25232310 24037379 25970245 25961798 24508170 30249741 15632085 25830018 21292972 21282665 25244985 20920257 23761445
EMBL
DQ105979
AAZ17653.1
NWSH01004992
PCG64517.1
AY154398
AAN52844.2
+ More
BABH01008498 BABH01008499 BABH01008500 ODYU01009490 SOQ53966.1 AGBW02011059 OWR47287.1 DQ105980 AAZ17654.1 KQ459984 KPJ19036.1 KP657644 AKL78869.1 RSAL01000201 RVE44440.1 KQ458793 KPJ05063.1 JTDY01000624 KOB76428.1 CH477638 EAT37969.1 GECL01000819 JAP05305.1 GBBI01002316 JAC16396.1 AB720744 BAM15639.1 GBGD01000916 JAC87973.1 GFTR01007359 JAW09067.1 NEVH01002992 PNF41019.1 JRES01001695 KNC20917.1 KQ981280 KYN43463.1 GL435030 EFN74191.1 ATLV01024553 KE525352 KFB51612.1 AXCN02000388 AXCN02000389 KQ976885 KYN07475.1 CH933808 EDW09490.2 ADTU01023469 ADTU01023470 ADTU01023471 KQ982911 KYQ49330.1 AAAB01008807 EAA04277.4 GL888066 EGI68379.1 KQ976511 KYM82529.1 KQ980824 KYN12561.1 GAMC01008426 JAB98129.1 CM000158 EDW91906.1 GAMC01008425 JAB98130.1 CH954179 EDV55741.1 CH940648 EDW62047.2 CH480816 EDW48158.1 CH916367 EDW01094.1 CM000362 CM002911 EDX07375.1 KMY94287.1 AE013599 AGB93558.1 AF260833 AF439752 AY051579 AAF76882.1 AAK93003.1 AAL32055.1 CCAG010001148 KQ434782 KZC04540.1 KK107154 EZA56881.1 QOIP01000001 RLU27541.1 GEBQ01006674 JAT33303.1 KZ288254 PBC30725.1 OUUW01000001 SPP73459.1 KQ435763 KOX75413.1 CH963849 EDW74016.1 GDHF01011983 JAI40331.1 AB370000 BAH22564.1 GDHF01029321 JAI22993.1 CM000071 EAL25019.2 CH479181 EDW31845.1 UFQS01001621 UFQT01001621 SSX11636.1 SSX31201.1 CP012524 ALC41674.1 CVRI01000043 CRK96201.1 GBXI01008487 JAD05805.1 GFXV01002857 MBW14662.1 GGMR01019255 MBY31874.1 GL732616 EFX70864.1 DS232104 EDS34926.1 ACPB03008045 GDIQ01216288 JAK35437.1 GDIP01007470 JAM96245.1 GL762910 EFZ20467.1 LNIX01000006 OXA52564.1 AXCM01001523 JXJN01007860 KQ761824 OAD56761.1 GDIQ01164868 JAK86857.1 APCN01000197 GDIQ01101821 JAL49905.1 ADMH02001280 ETN63223.1 KK113777 KFM61014.1 CH920905 EDW04524.1 KA649196 AFP63825.1 GDIP01159633 JAJ63769.1 LRGB01002836 KZS06180.1 GECU01031913 JAS75793.1 GGLE01005702 MBY09828.1
BABH01008498 BABH01008499 BABH01008500 ODYU01009490 SOQ53966.1 AGBW02011059 OWR47287.1 DQ105980 AAZ17654.1 KQ459984 KPJ19036.1 KP657644 AKL78869.1 RSAL01000201 RVE44440.1 KQ458793 KPJ05063.1 JTDY01000624 KOB76428.1 CH477638 EAT37969.1 GECL01000819 JAP05305.1 GBBI01002316 JAC16396.1 AB720744 BAM15639.1 GBGD01000916 JAC87973.1 GFTR01007359 JAW09067.1 NEVH01002992 PNF41019.1 JRES01001695 KNC20917.1 KQ981280 KYN43463.1 GL435030 EFN74191.1 ATLV01024553 KE525352 KFB51612.1 AXCN02000388 AXCN02000389 KQ976885 KYN07475.1 CH933808 EDW09490.2 ADTU01023469 ADTU01023470 ADTU01023471 KQ982911 KYQ49330.1 AAAB01008807 EAA04277.4 GL888066 EGI68379.1 KQ976511 KYM82529.1 KQ980824 KYN12561.1 GAMC01008426 JAB98129.1 CM000158 EDW91906.1 GAMC01008425 JAB98130.1 CH954179 EDV55741.1 CH940648 EDW62047.2 CH480816 EDW48158.1 CH916367 EDW01094.1 CM000362 CM002911 EDX07375.1 KMY94287.1 AE013599 AGB93558.1 AF260833 AF439752 AY051579 AAF76882.1 AAK93003.1 AAL32055.1 CCAG010001148 KQ434782 KZC04540.1 KK107154 EZA56881.1 QOIP01000001 RLU27541.1 GEBQ01006674 JAT33303.1 KZ288254 PBC30725.1 OUUW01000001 SPP73459.1 KQ435763 KOX75413.1 CH963849 EDW74016.1 GDHF01011983 JAI40331.1 AB370000 BAH22564.1 GDHF01029321 JAI22993.1 CM000071 EAL25019.2 CH479181 EDW31845.1 UFQS01001621 UFQT01001621 SSX11636.1 SSX31201.1 CP012524 ALC41674.1 CVRI01000043 CRK96201.1 GBXI01008487 JAD05805.1 GFXV01002857 MBW14662.1 GGMR01019255 MBY31874.1 GL732616 EFX70864.1 DS232104 EDS34926.1 ACPB03008045 GDIQ01216288 JAK35437.1 GDIP01007470 JAM96245.1 GL762910 EFZ20467.1 LNIX01000006 OXA52564.1 AXCM01001523 JXJN01007860 KQ761824 OAD56761.1 GDIQ01164868 JAK86857.1 APCN01000197 GDIQ01101821 JAL49905.1 ADMH02001280 ETN63223.1 KK113777 KFM61014.1 CH920905 EDW04524.1 KA649196 AFP63825.1 GDIP01159633 JAJ63769.1 LRGB01002836 KZS06180.1 GECU01031913 JAS75793.1 GGLE01005702 MBY09828.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053240
UP000283053
UP000053268
+ More
UP000037510 UP000008820 UP000095301 UP000235965 UP000037069 UP000075880 UP000095300 UP000078541 UP000000311 UP000030765 UP000075886 UP000078542 UP000009192 UP000005205 UP000075809 UP000075903 UP000007062 UP000007755 UP000078540 UP000091820 UP000078492 UP000002282 UP000008711 UP000008792 UP000001292 UP000001070 UP000000304 UP000000803 UP000092444 UP000076502 UP000053097 UP000279307 UP000192221 UP000242457 UP000268350 UP000053105 UP000005203 UP000007798 UP000001819 UP000008744 UP000092553 UP000183832 UP000000305 UP000002320 UP000015103 UP000075902 UP000069272 UP000075885 UP000198287 UP000075884 UP000076408 UP000075883 UP000075920 UP000092460 UP000075840 UP000075881 UP000000673 UP000054359 UP000075900 UP000076858 UP000075901
UP000037510 UP000008820 UP000095301 UP000235965 UP000037069 UP000075880 UP000095300 UP000078541 UP000000311 UP000030765 UP000075886 UP000078542 UP000009192 UP000005205 UP000075809 UP000075903 UP000007062 UP000007755 UP000078540 UP000091820 UP000078492 UP000002282 UP000008711 UP000008792 UP000001292 UP000001070 UP000000304 UP000000803 UP000092444 UP000076502 UP000053097 UP000279307 UP000192221 UP000242457 UP000268350 UP000053105 UP000005203 UP000007798 UP000001819 UP000008744 UP000092553 UP000183832 UP000000305 UP000002320 UP000015103 UP000075902 UP000069272 UP000075885 UP000198287 UP000075884 UP000076408 UP000075883 UP000075920 UP000092460 UP000075840 UP000075881 UP000000673 UP000054359 UP000075900 UP000076858 UP000075901
Pfam
PF00209 SNF
SUPFAM
SSF161070
SSF161070
ProteinModelPortal
Q45VA3
A0A2A4IX52
Q86MC4
H9JAS4
A0A2H1WLW5
A0A212F0P2
+ More
Q45VA2 A0A194RSJ2 A0A0G3VG04 A0A3S2NTV6 A0A194QHP4 A0A0L7LLJ2 Q16TV9 A0A1I8MCE3 A0A0V0GBJ2 A0A023F4F4 I2FKF4 A0A069DV82 A0A224X9N0 A0A2J7RJL9 A0A0L0BLL6 A0A182IX90 A0A1I8PFW0 A0A195FSP1 E1ZXA1 A0A084WN18 A0A182QPG6 A0A195D3J0 B4KSI5 A0A158NQQ9 A0A151WND3 A0A182UWK2 Q7QJ94 F4WBV1 A0A195BE92 A0A1A9WR92 A0A195DI93 W8C6P2 B4P656 W8BA44 B3NPH4 B4LMF2 B4HSZ4 B4J7H0 B4QI04 A0A0B4KEX2 Q7K4Y6 A0A1B0FKI7 A0A154P078 A0A026WMI7 A0A3L8E4C5 A0A1W4UKV4 A0A1B6MBJ9 A0A2A3EG39 A0A3B0JJ43 A0A0M9A3V3 A0A087ZQ28 B4MPH7 A0A0K8VN76 B9A0V7 A0A0K8U8S5 Q291U3 B4GA93 A0A336MNN5 A0A0M5IXL3 A0A1J1ICN7 A0A0A1X506 A0A2H8TLX0 A0A2S2PRA9 E9HBL2 B0WUE7 T1I1H1 A0A0P5IDE2 A0A0P6BYM5 E9IG58 A0A182UCW0 A0A182FDF5 A0A182PRE6 A0A226E682 A0A182NI32 A0A182XYN8 A0A182MIW2 A0A182WJF0 A0A1B0B3H9 A0A310SJT6 A0A0P5LUT0 A0A182I335 A0A0P5RJ52 A0A182X3Y4 A0A182JRM0 W5JIV4 A0A087T7C5 A0A182R7A4 B4K361 T1PKA4 A0A0N8ACY8 A0A164NRL7 A0A1B6HM80 A0A182SRQ8 A0A2R5LK72
Q45VA2 A0A194RSJ2 A0A0G3VG04 A0A3S2NTV6 A0A194QHP4 A0A0L7LLJ2 Q16TV9 A0A1I8MCE3 A0A0V0GBJ2 A0A023F4F4 I2FKF4 A0A069DV82 A0A224X9N0 A0A2J7RJL9 A0A0L0BLL6 A0A182IX90 A0A1I8PFW0 A0A195FSP1 E1ZXA1 A0A084WN18 A0A182QPG6 A0A195D3J0 B4KSI5 A0A158NQQ9 A0A151WND3 A0A182UWK2 Q7QJ94 F4WBV1 A0A195BE92 A0A1A9WR92 A0A195DI93 W8C6P2 B4P656 W8BA44 B3NPH4 B4LMF2 B4HSZ4 B4J7H0 B4QI04 A0A0B4KEX2 Q7K4Y6 A0A1B0FKI7 A0A154P078 A0A026WMI7 A0A3L8E4C5 A0A1W4UKV4 A0A1B6MBJ9 A0A2A3EG39 A0A3B0JJ43 A0A0M9A3V3 A0A087ZQ28 B4MPH7 A0A0K8VN76 B9A0V7 A0A0K8U8S5 Q291U3 B4GA93 A0A336MNN5 A0A0M5IXL3 A0A1J1ICN7 A0A0A1X506 A0A2H8TLX0 A0A2S2PRA9 E9HBL2 B0WUE7 T1I1H1 A0A0P5IDE2 A0A0P6BYM5 E9IG58 A0A182UCW0 A0A182FDF5 A0A182PRE6 A0A226E682 A0A182NI32 A0A182XYN8 A0A182MIW2 A0A182WJF0 A0A1B0B3H9 A0A310SJT6 A0A0P5LUT0 A0A182I335 A0A0P5RJ52 A0A182X3Y4 A0A182JRM0 W5JIV4 A0A087T7C5 A0A182R7A4 B4K361 T1PKA4 A0A0N8ACY8 A0A164NRL7 A0A1B6HM80 A0A182SRQ8 A0A2R5LK72
PDB
4XPA
E-value=0,
Score=2216
Ontologies
GO
GO:0016021
GO:0005328
GO:0015293
GO:0015874
GO:0005330
GO:0042734
GO:0032809
GO:0051583
GO:0043005
GO:0005886
GO:0099509
GO:0030431
GO:0008344
GO:0046872
GO:1990834
GO:0042745
GO:0098793
GO:0019811
GO:0005887
GO:0015872
GO:0006836
GO:0006468
GO:0005524
GO:0031683
GO:0001664
GO:0005525
GO:0007264
GO:0003824
PANTHER
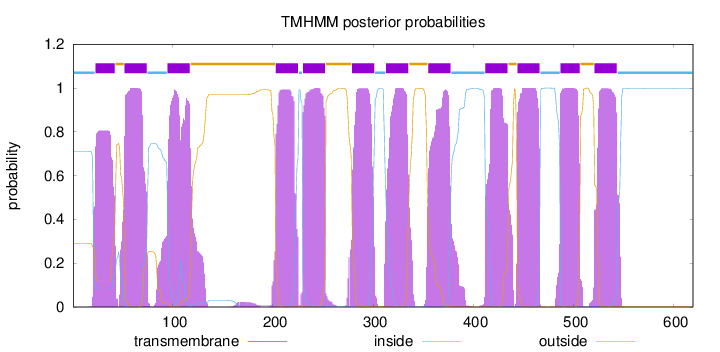
Topology
Subcellular location
Cell membrane
Length:
619
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
266.25967
Exp number, first 60 AAs:
26.62077
Total prob of N-in:
0.71042
POSSIBLE N-term signal
sequence
inside
1 - 22
TMhelix
23 - 42
outside
43 - 51
TMhelix
52 - 74
inside
75 - 94
TMhelix
95 - 117
outside
118 - 202
TMhelix
203 - 225
inside
226 - 229
TMhelix
230 - 252
outside
253 - 278
TMhelix
279 - 301
inside
302 - 312
TMhelix
313 - 335
outside
336 - 354
TMhelix
355 - 377
inside
378 - 411
TMhelix
412 - 434
outside
435 - 443
TMhelix
444 - 466
inside
467 - 486
TMhelix
487 - 506
outside
507 - 520
TMhelix
521 - 543
inside
544 - 619
Population Genetic Test Statistics
Pi
203.655259
Theta
171.773883
Tajima's D
0.897598
CLR
0.350999
CSRT
0.630468476576171
Interpretation
Uncertain
