Gene
KWMTBOMO05498 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006623
Annotation
PREDICTED:_C-type_lectin_11_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.317
Sequence
CDS
ATGAAGAGTGCTTTTATTTGTTTCTGCTTTTTGGCGATTGCCAGTGCCCCGCCTCCGAGTGTTTCGAAGCAATACCGCTCCGACTACGTATATAATAAAGACACCAACGCGTTTTACAAACTACACACCGACAGCGCTAAGATATGGGACGCCAAGAGCTCTTGCACCACTGAGGGAGCCCAGTTGATGGTGCCGGCTTCAGAACAGGATATAATTCAGCTTCATTCGATGTTCAAGAGATTTCCAGATCTGGGGAACTATGTTTGGGTGGATGAAGACGGAAAAGACCATGAATCTGCAGAGGAACAGCCGATGATTGATTTGTCAGACTCGGTAACGGAAGCGATGAGGTCTAGGTTCGCATTACAAGGGTGCGATGTGGTTACTCGGCAGGGAGAAATCGAAACCTCACCCTGCTACAACCGATTACCTTTCATTTGCAAAGTGGAAGCCATTGATGCGCCTTACGATACGCACTGTGGGGTTTTTGCTATAGGGTACCAATACGTGGAGAGCACTGGAAGCTGCTATAAGATCTCCAAAGTAGCCTATTCCTGGAACCAAGCTTATGACGAATGTCAAGCCGAGAATGCCCACCTGGTGGTCCTTAATTCGGAAGCGGAGATGTTGGTCGTGAAGAATTTGACGAACGCAGCGGCCCCAGTTGACGAAGCCCAGACCACATATTTCTTTTATGCCGGTTTCAGAGCACAGGAACCCACTAAAAACGAAACTCCCGTGTTCAAGACGATTTTCAATCAAACATTACAAGAAGCCGGTTTCAGTGGCTGGTCCGAAAACGAACCTAACAACGCGAACAACATCGAATTTTGCGGCACGCTCTTCAAAAACGACGGCAAACTGAATGACGTCGACTGCTCCGATAAATACGCCTTCATATGCGAAAAGGAGGTGCATATGTAA
Protein
MKSAFICFCFLAIASAPPPSVSKQYRSDYVYNKDTNAFYKLHTDSAKIWDAKSSCTTEGAQLMVPASEQDIIQLHSMFKRFPDLGNYVWVDEDGKDHESAEEQPMIDLSDSVTEAMRSRFALQGCDVVTRQGEIETSPCYNRLPFICKVEAIDAPYDTHCGVFAIGYQYVESTGSCYKISKVAYSWNQAYDECQAENAHLVVLNSEAEMLVVKNLTNAAAPVDEAQTTYFFYAGFRAQEPTKNETPVFKTIFNQTLQEAGFSGWSENEPNNANNIEFCGTLFKNDGKLNDVDCSDKYAFICEKEVHM
Summary
Uniprot
Q7Z1E5
H9JAS8
D3YFC2
O76301
H9AA74
A0A2H1VX16
+ More
A0A0H4T1R3 A0A2A4J067 I1VR02 A0A3S2LDL7 A0A0L7KZ71 A0A194QNJ2 A0A194RNF5 S4PJZ4 A0A1C9EGM6 A0A219YXH3 A0A219YXH1 A0A212EQ02 A0A219YXG9 A0A096ZGV2 A0A219YXG6 A0A194QAQ3 A0A212FF49 A0A194QMK1 I4DJJ2 A0A0N1IPT5 G5CJE6 A0A194PEZ5 A0A219YXK5 A0A1E1WP12 A0A219YXI5 Q5UAW7 A0A219YXG7 A0A3G1HH41 B1A5Z5 A0A3G1HH66 A0A219YXG8 A0A3S2P588 H9IYF4 A0A194QLC9 C0L7M1 A0A219YXH0 A0A3S2TBA6 A0A096ZGV0 R9UPV0 A0A0D3M5W6 A0A2A4K0G1 O97363 Q9NBV9 A0A096ZGV4 A0A0N1PJ18 A0A219YXH8 A0A219YXI8 I1VR03 B3IYT1 A0A3S2NP58 A0A219YXH7 A0A219YXI3 A0A1E1WQ86 Q5UAW8 Q0KKW8 A0A194QAQ8 D2X2F6 A0A2W1BVD9 A0A2W1BTF0 A0A219YXH4 B7SVM1 A0A219YXL1 A0A219YXI4 C0L7M0 A0A0L7LF02 A0A194QKT8 A0A194RL20 A0A194PE23 A0A3S2LAT9 A0A3S2L865 A0A096ZGV1 A0A219YXH2 A0A2H1WK53 A0A219YXH6 A0A194PUI0 A0A0L7LUC3 A0A219YXH5 A0A173M1W5 A0A2A4IYF0 I1VR05 A0A0L7LRB5 A0A3S2NK24 A0A2W1BVG4 Q19AB1 A0A2H1VQU1 A0A2A4J3T7 A0A2A4J242 A0A3S2TGZ3 A0A194QHR4 A0A2W1BYL2 Q5MGF0
A0A0H4T1R3 A0A2A4J067 I1VR02 A0A3S2LDL7 A0A0L7KZ71 A0A194QNJ2 A0A194RNF5 S4PJZ4 A0A1C9EGM6 A0A219YXH3 A0A219YXH1 A0A212EQ02 A0A219YXG9 A0A096ZGV2 A0A219YXG6 A0A194QAQ3 A0A212FF49 A0A194QMK1 I4DJJ2 A0A0N1IPT5 G5CJE6 A0A194PEZ5 A0A219YXK5 A0A1E1WP12 A0A219YXI5 Q5UAW7 A0A219YXG7 A0A3G1HH41 B1A5Z5 A0A3G1HH66 A0A219YXG8 A0A3S2P588 H9IYF4 A0A194QLC9 C0L7M1 A0A219YXH0 A0A3S2TBA6 A0A096ZGV0 R9UPV0 A0A0D3M5W6 A0A2A4K0G1 O97363 Q9NBV9 A0A096ZGV4 A0A0N1PJ18 A0A219YXH8 A0A219YXI8 I1VR03 B3IYT1 A0A3S2NP58 A0A219YXH7 A0A219YXI3 A0A1E1WQ86 Q5UAW8 Q0KKW8 A0A194QAQ8 D2X2F6 A0A2W1BVD9 A0A2W1BTF0 A0A219YXH4 B7SVM1 A0A219YXL1 A0A219YXI4 C0L7M0 A0A0L7LF02 A0A194QKT8 A0A194RL20 A0A194PE23 A0A3S2LAT9 A0A3S2L865 A0A096ZGV1 A0A219YXH2 A0A2H1WK53 A0A219YXH6 A0A194PUI0 A0A0L7LUC3 A0A219YXH5 A0A173M1W5 A0A2A4IYF0 I1VR05 A0A0L7LRB5 A0A3S2NK24 A0A2W1BVG4 Q19AB1 A0A2H1VQU1 A0A2A4J3T7 A0A2A4J242 A0A3S2TGZ3 A0A194QHR4 A0A2W1BYL2 Q5MGF0
Pubmed
EMBL
AY297159
AAP50846.1
BABH01008479
BABH01008480
BABH01008481
BABH01008482
+ More
GU454799 ADD13530.1 AF053131 AAC33576.1 JN880426 AFC35299.1 ODYU01004970 SOQ45379.1 KP406777 AKP99431.1 NWSH01004470 PCG65068.1 JQ250828 AFI47448.1 RSAL01000201 RVE44445.1 JTDY01004215 KOB68445.1 KQ458793 KPJ05056.1 KQ459984 KPJ19042.1 GAIX01004755 JAA87805.1 KX778610 AON96650.1 KX550128 AQY54445.1 KX550126 AQY54443.1 AGBW02013342 OWR43573.1 KX550120 AQY54437.1 KF752420 AIR95999.1 KX550121 AQY54438.1 KQ459232 KPJ02552.1 AGBW02008849 OWR52366.1 KQ461198 KPJ06185.1 AK401460 BAM18082.1 KQ460044 KPJ18317.1 JN133501 AEO52696.1 KQ459606 KPI91593.1 KX550129 AQY54446.1 GDQN01002397 JAT88657.1 KX550141 AQY54458.1 AY768812 AAV41237.2 KX550123 AQY54440.1 KY111311 AQX37247.1 EU426550 ABZ81710.1 KY111312 AQX37248.1 KX550122 AQY54439.1 RSAL01000019 RVE52735.1 BABH01020449 KPJ06184.1 FJ666211 ACN41859.1 KX550127 AQY54444.1 RSAL01001098 RVE40947.1 KF752419 AIR95998.1 KC733761 AGN70857.1 KF285999 AIC33761.1 NWSH01000351 PCG77132.1 AJ011573 CAB38429.1 AF242202 AAF91316.2 KF752421 AIR96000.1 KPJ18324.1 KX550124 AQY54441.1 KX550142 AQY54459.1 JQ269654 AFI47449.1 AB444137 BAG54784.1 RVE52734.1 KX550133 AQY54450.1 KX550134 AQY54451.1 GDQN01002063 JAT88991.1 AY768811 AAV41236.1 AB219148 BAF03496.1 KPJ02557.1 GQ487489 ADB12587.1 KZ149926 PZC77594.1 KZ149905 PZC78329.1 KX550130 AQY54447.1 EU770390 ACI32834.1 KX550139 AQY54456.1 KX550138 AQY54455.1 FJ666210 ACN41858.1 JTDY01001404 KOB73959.1 KPJ06183.1 KQ460207 KPJ16676.1 KPI91591.1 RSAL01000062 RVE49558.1 RSAL01000824 RVE41129.1 KF752422 AIR96001.1 KX550125 AQY54442.1 ODYU01009180 SOQ53429.1 KX550132 AQY54449.1 KQ459597 KPI94795.1 JTDY01000107 KOB78821.1 KX550131 AQY54448.1 LC150882 BAV01248.1 NWSH01005098 PCG64438.1 JQ269656 AFI47451.1 JTDY01000269 KOB77990.1 RVE49559.1 PZC78331.1 DQ533877 ABF83203.1 ODYU01003868 SOQ43168.1 NWSH01003594 PCG66072.1 PCG66071.1 RSAL01000150 RVE45770.1 KPJ05052.1 PZC78327.1 AY829836 AAV91450.1
GU454799 ADD13530.1 AF053131 AAC33576.1 JN880426 AFC35299.1 ODYU01004970 SOQ45379.1 KP406777 AKP99431.1 NWSH01004470 PCG65068.1 JQ250828 AFI47448.1 RSAL01000201 RVE44445.1 JTDY01004215 KOB68445.1 KQ458793 KPJ05056.1 KQ459984 KPJ19042.1 GAIX01004755 JAA87805.1 KX778610 AON96650.1 KX550128 AQY54445.1 KX550126 AQY54443.1 AGBW02013342 OWR43573.1 KX550120 AQY54437.1 KF752420 AIR95999.1 KX550121 AQY54438.1 KQ459232 KPJ02552.1 AGBW02008849 OWR52366.1 KQ461198 KPJ06185.1 AK401460 BAM18082.1 KQ460044 KPJ18317.1 JN133501 AEO52696.1 KQ459606 KPI91593.1 KX550129 AQY54446.1 GDQN01002397 JAT88657.1 KX550141 AQY54458.1 AY768812 AAV41237.2 KX550123 AQY54440.1 KY111311 AQX37247.1 EU426550 ABZ81710.1 KY111312 AQX37248.1 KX550122 AQY54439.1 RSAL01000019 RVE52735.1 BABH01020449 KPJ06184.1 FJ666211 ACN41859.1 KX550127 AQY54444.1 RSAL01001098 RVE40947.1 KF752419 AIR95998.1 KC733761 AGN70857.1 KF285999 AIC33761.1 NWSH01000351 PCG77132.1 AJ011573 CAB38429.1 AF242202 AAF91316.2 KF752421 AIR96000.1 KPJ18324.1 KX550124 AQY54441.1 KX550142 AQY54459.1 JQ269654 AFI47449.1 AB444137 BAG54784.1 RVE52734.1 KX550133 AQY54450.1 KX550134 AQY54451.1 GDQN01002063 JAT88991.1 AY768811 AAV41236.1 AB219148 BAF03496.1 KPJ02557.1 GQ487489 ADB12587.1 KZ149926 PZC77594.1 KZ149905 PZC78329.1 KX550130 AQY54447.1 EU770390 ACI32834.1 KX550139 AQY54456.1 KX550138 AQY54455.1 FJ666210 ACN41858.1 JTDY01001404 KOB73959.1 KPJ06183.1 KQ460207 KPJ16676.1 KPI91591.1 RSAL01000062 RVE49558.1 RSAL01000824 RVE41129.1 KF752422 AIR96001.1 KX550125 AQY54442.1 ODYU01009180 SOQ53429.1 KX550132 AQY54449.1 KQ459597 KPI94795.1 JTDY01000107 KOB78821.1 KX550131 AQY54448.1 LC150882 BAV01248.1 NWSH01005098 PCG64438.1 JQ269656 AFI47451.1 JTDY01000269 KOB77990.1 RVE49559.1 PZC78331.1 DQ533877 ABF83203.1 ODYU01003868 SOQ43168.1 NWSH01003594 PCG66072.1 PCG66071.1 RSAL01000150 RVE45770.1 KPJ05052.1 PZC78327.1 AY829836 AAV91450.1
Proteomes
Interpro
IPR001304
C-type_lectin-like
+ More
IPR016186 C-type_lectin-like/link_sf
IPR016187 CTDL_fold
IPR018378 C-type_lectin_CS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR003563 OxG-triPHTase
IPR000086 NUDIX_hydrolase_dom
IPR036179 Ig-like_dom_sf
IPR013783 Ig-like_fold
IPR016186 C-type_lectin-like/link_sf
IPR016187 CTDL_fold
IPR018378 C-type_lectin_CS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR003563 OxG-triPHTase
IPR000086 NUDIX_hydrolase_dom
IPR036179 Ig-like_dom_sf
IPR013783 Ig-like_fold
Gene 3D
ProteinModelPortal
Q7Z1E5
H9JAS8
D3YFC2
O76301
H9AA74
A0A2H1VX16
+ More
A0A0H4T1R3 A0A2A4J067 I1VR02 A0A3S2LDL7 A0A0L7KZ71 A0A194QNJ2 A0A194RNF5 S4PJZ4 A0A1C9EGM6 A0A219YXH3 A0A219YXH1 A0A212EQ02 A0A219YXG9 A0A096ZGV2 A0A219YXG6 A0A194QAQ3 A0A212FF49 A0A194QMK1 I4DJJ2 A0A0N1IPT5 G5CJE6 A0A194PEZ5 A0A219YXK5 A0A1E1WP12 A0A219YXI5 Q5UAW7 A0A219YXG7 A0A3G1HH41 B1A5Z5 A0A3G1HH66 A0A219YXG8 A0A3S2P588 H9IYF4 A0A194QLC9 C0L7M1 A0A219YXH0 A0A3S2TBA6 A0A096ZGV0 R9UPV0 A0A0D3M5W6 A0A2A4K0G1 O97363 Q9NBV9 A0A096ZGV4 A0A0N1PJ18 A0A219YXH8 A0A219YXI8 I1VR03 B3IYT1 A0A3S2NP58 A0A219YXH7 A0A219YXI3 A0A1E1WQ86 Q5UAW8 Q0KKW8 A0A194QAQ8 D2X2F6 A0A2W1BVD9 A0A2W1BTF0 A0A219YXH4 B7SVM1 A0A219YXL1 A0A219YXI4 C0L7M0 A0A0L7LF02 A0A194QKT8 A0A194RL20 A0A194PE23 A0A3S2LAT9 A0A3S2L865 A0A096ZGV1 A0A219YXH2 A0A2H1WK53 A0A219YXH6 A0A194PUI0 A0A0L7LUC3 A0A219YXH5 A0A173M1W5 A0A2A4IYF0 I1VR05 A0A0L7LRB5 A0A3S2NK24 A0A2W1BVG4 Q19AB1 A0A2H1VQU1 A0A2A4J3T7 A0A2A4J242 A0A3S2TGZ3 A0A194QHR4 A0A2W1BYL2 Q5MGF0
A0A0H4T1R3 A0A2A4J067 I1VR02 A0A3S2LDL7 A0A0L7KZ71 A0A194QNJ2 A0A194RNF5 S4PJZ4 A0A1C9EGM6 A0A219YXH3 A0A219YXH1 A0A212EQ02 A0A219YXG9 A0A096ZGV2 A0A219YXG6 A0A194QAQ3 A0A212FF49 A0A194QMK1 I4DJJ2 A0A0N1IPT5 G5CJE6 A0A194PEZ5 A0A219YXK5 A0A1E1WP12 A0A219YXI5 Q5UAW7 A0A219YXG7 A0A3G1HH41 B1A5Z5 A0A3G1HH66 A0A219YXG8 A0A3S2P588 H9IYF4 A0A194QLC9 C0L7M1 A0A219YXH0 A0A3S2TBA6 A0A096ZGV0 R9UPV0 A0A0D3M5W6 A0A2A4K0G1 O97363 Q9NBV9 A0A096ZGV4 A0A0N1PJ18 A0A219YXH8 A0A219YXI8 I1VR03 B3IYT1 A0A3S2NP58 A0A219YXH7 A0A219YXI3 A0A1E1WQ86 Q5UAW8 Q0KKW8 A0A194QAQ8 D2X2F6 A0A2W1BVD9 A0A2W1BTF0 A0A219YXH4 B7SVM1 A0A219YXL1 A0A219YXI4 C0L7M0 A0A0L7LF02 A0A194QKT8 A0A194RL20 A0A194PE23 A0A3S2LAT9 A0A3S2L865 A0A096ZGV1 A0A219YXH2 A0A2H1WK53 A0A219YXH6 A0A194PUI0 A0A0L7LUC3 A0A219YXH5 A0A173M1W5 A0A2A4IYF0 I1VR05 A0A0L7LRB5 A0A3S2NK24 A0A2W1BVG4 Q19AB1 A0A2H1VQU1 A0A2A4J3T7 A0A2A4J242 A0A3S2TGZ3 A0A194QHR4 A0A2W1BYL2 Q5MGF0
PDB
1DV8
E-value=9.40729e-06,
Score=116
Ontologies
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.992019
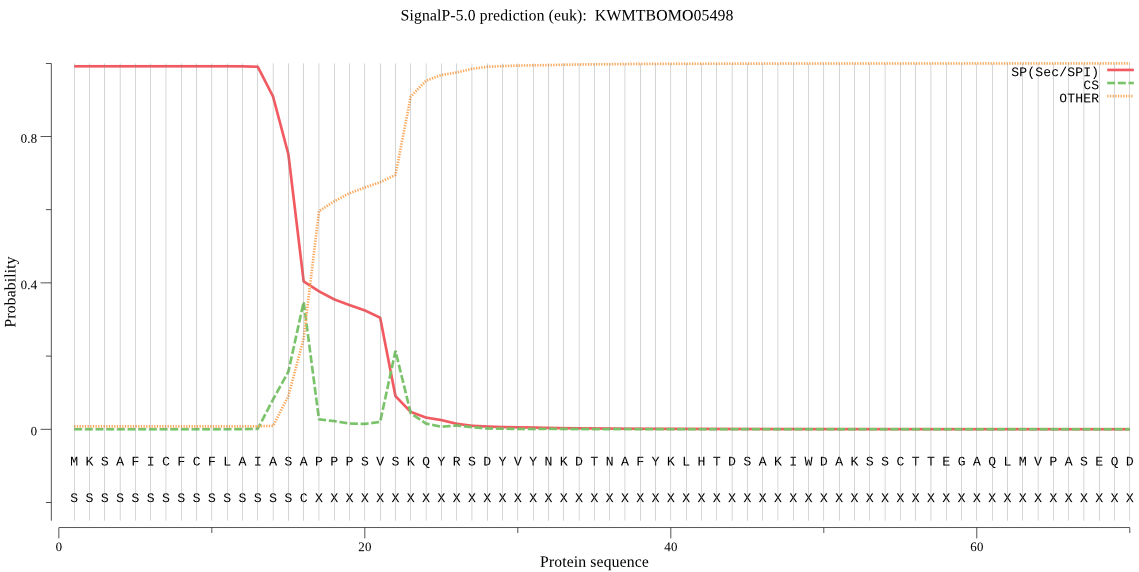
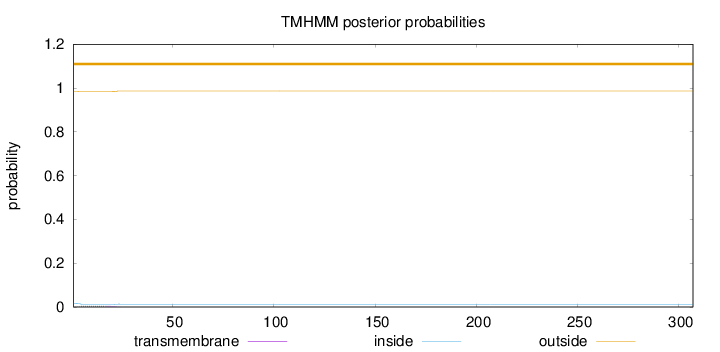
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0896599999999999
Exp number, first 60 AAs:
0.08935
Total prob of N-in:
0.01607
outside
1 - 307
Population Genetic Test Statistics
Pi
185.027433
Theta
173.56001
Tajima's D
0.130485
CLR
0.773445
CSRT
0.409429528523574
Interpretation
Uncertain
