Gene
KWMTBOMO05483
Pre Gene Modal
BGIBMGA006627
Annotation
PREDICTED:_RNA_polymerase_I-specific_transcription_initiation_factor_RRN3_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.478 PlasmaMembrane Reliability : 1.815
Sequence
CDS
ATGTCGAGAGAAGGTTATAGCCCAGAAAAGAAAGAAAGTGACAACAAGGCTGTTTTAGAGCGATTACAGTCTTATTTGAAAGACAAGAAAAACACTGACGTTTCCGACACGAGCGCCGGGTACAGCAAACCAAGTACGAGCTACGATCCGAACACAAATACTGTCCCGAACATAGAAGATAGTAGCAATCACGAAAGACTTGAAGAAGTAATGGCGGTAGCGGCTAGGAAAAGCGTCGCGTCCTTGATGCAAAAAACTCTGGATCGCCAAGCGGCTTTAGCTCGTACTAGATTGAAATTCTCCGAAAAACAAATAGAACAGATACTTCTTAATTATGCTAAAACAGGAAACATTGGACCTTACTCGGAATTAGTCAGAGTTTTGAGAGATTACCCATTAAATGATGAGAATTTTCGGATTATTCTCGAAGATTGTCTCTCTTGCGTTCTGATTATGGGACAGGATTTCAAGCAGTTTGTAGAAGCAGTGTGCAATGTTGAGTGGGCTAGTAGGTGTGAAAAGTTAGTCAACCTTTTCAGCACCTTCATCATTAGCCTAGTCACTGCACATATTTACCATTGTCCGCAAGTGATGAGCTGTCTGGTTAAGGTTTTTAAAGCTAATGAAGAAAACTGGGATGACACTCCACCAACAGAGAAATTGAATAGCTGGTCTAACATTCATACAATCATAGGACAAATTATCGCAATAATGCCGATGACAAATGAACTTTTAATGGAGACTGTGATAGATCAATTTCCATATCACAAAGCTGGTTGTAGTTCAAATAGATCTTTCATATATAACCTTATATGGATTAGCAAATACCTACCTTCACTTACAGAGGAAATTATGATGATTATTATTAATAGAATGATAGTAATGGATGGCAATATAGTGGAAAGTGACAATAAAATGAAACAGAATGACACACTATTTGACATGGATGTGGAACAACATGATGACATCTCTGAAACCCTAGACTATAGTATGCTAGAAGTATTGCACTGGCTACAGGAAGAAAGAGACCCTATTTTGCACCACATGTGCACAGCTTTTGAAAAAATAATACTACCAACTTACGGAATCAGGCATGTTCAATTTTTACTGCTGTATACAATATCAATAAACCAACATTGCGCTGACCGAGTTCTCAACAATCTGTGGACAGTAGCGGCAGGGCTTCAAGGTCTAGGACCTGGTGCATTGGCAACAAGACGAACAGCTGCCAGTCATCTTGCAGGCCTGCTGGCTCGTTGTGTTCGAGTTCCTAACTCAAGATTAGTACAATTCTTGAAGACAATGACAGATTGGTGCCATTCCTACATCTCTGCAGCTCAAGAGTCCACCACTGTTACTGATAACATTAAAGCTCATGGAGCCTTCCATGCTATATGTCATGCTGTGTTTTACCTAATTGCATTCAAACACCAGCATCTGTTTATGAGTAAAGAAAGCTTAAACTTTGTGGAGTCTCTTAATCTTCCTCGGCTAGTGGCTTGCACACTGAATCCTCTCCGCACCTGCCCACCGCAAGTGACACGAGCATTTTCATCGGTGACTCGATCTCACCAAATTGTGTATTGTCAAGCTATTATTGAGAACAACGCGCGACACACGTTACAGTCTGCAGATATTTTGCAGTATGATGAATGGTTTCCATACGACCCTTATCAATTACCTAGAAGCGGTAAAATAATATGGCCGCTGTGCATCGATTACAAAGACTGGTTAAAAGAAGGCGATGACGAAACTCAATACTCAAACATGAAAAAGAAAATGGAGGCAGACGATGACGACTTTTTAATGTCTGTTAGTCCATGTTCTAAACTAGCAAGCTCACTGTCCAATGGAGTATTACCTGGATTTAGAACAAGTGAAAGCATACCATACTAA
Protein
MSREGYSPEKKESDNKAVLERLQSYLKDKKNTDVSDTSAGYSKPSTSYDPNTNTVPNIEDSSNHERLEEVMAVAARKSVASLMQKTLDRQAALARTRLKFSEKQIEQILLNYAKTGNIGPYSELVRVLRDYPLNDENFRIILEDCLSCVLIMGQDFKQFVEAVCNVEWASRCEKLVNLFSTFIISLVTAHIYHCPQVMSCLVKVFKANEENWDDTPPTEKLNSWSNIHTIIGQIIAIMPMTNELLMETVIDQFPYHKAGCSSNRSFIYNLIWISKYLPSLTEEIMMIIINRMIVMDGNIVESDNKMKQNDTLFDMDVEQHDDISETLDYSMLEVLHWLQEERDPILHHMCTAFEKIILPTYGIRHVQFLLLYTISINQHCADRVLNNLWTVAAGLQGLGPGALATRRTAASHLAGLLARCVRVPNSRLVQFLKTMTDWCHSYISAAQESTTVTDNIKAHGAFHAICHAVFYLIAFKHQHLFMSKESLNFVESLNLPRLVACTLNPLRTCPPQVTRAFSSVTRSHQIVYCQAIIENNARHTLQSADILQYDEWFPYDPYQLPRSGKIIWPLCIDYKDWLKEGDDETQYSNMKKKMEADDDDFLMSVSPCSKLASSLSNGVLPGFRTSESIPY
Summary
Uniprot
H9JAT2
A0A2A4JQC3
A0A2H1WT02
S4P8A1
A0A194QHM7
A0A194RSK7
+ More
A0A1W4X9H0 A0A0C9RYU6 D6X392 A0A1Y1MD29 J3JXX8 N6U031 A0A146LTP3 A0A0A9Z8Y3 A0A182ILW1 A0A0K8TBF4 A0A2J7RRA3 A0A1Q3EXL6 A0A182Q1R4 Q16ML1 A0A2M4BHG3 A0A2M4BI58 A0A182F423 W5JSQ5 A0A2S2Q1X8 A0A2M4BHI1 A0A0P4VPA0 A0A084VKL4 A0A182NBD8 A0A1L8DFQ4 A0A1L8DFP2 B0WU55 A0A2M4AG85 K7IVM8 A0A069DV56 A0A224X9U2 A0A232EQM4 A0A2M3Z3M2 A0A182JV57 A0A1B0CNP2 A0A182VU30 A0A182GHK7 A0A023EUD2 A0A1J1IZU4 A0A0V0G315 E0VUA1 A0A1A8GYV7 A0A1S3JXZ1 A0A182YFR3 A0A336MUU1 U5EH15 A0A067QXA9 A0A3B5LSG5 A0A182RLD3 A0A023F0Q3 J9JZJ3 A0A182LLX0 A0A1L8DFN8 M4A6Q4 A0A1S4GZW1 A0A1A8A9N0 A0A1A9YPW0 A0A182LU10 A0A1B0C529 B4GXB4 A0A2J7RRA4 A0A1A8BUZ5 A0A1A9UWV3 A0A1B0G3J9 A0A3Q2CDM5 Q29K22 J9JUB4 A0A182PED6 A0A1A8KEG0 A0A2H8TZD6 A0A182V710 A0A1A9ZW01 A0A0K8TL75 A0A0L0C3Y4 A0A3P8Z3Q6 A0A1A8QLR3 A0A1A8LYB4 A0A154P9K5 A0A1A7YLF6 A0A034V3T8 A0A0M4E7D0 A0A3Q2QID6 A0A146V7Q4 A0A3P9P4G8 A0A3B0JTV3 B4LQQ2 A0A0A1X2P8 Q4SEX9 A0A1S3R6B4 G3NKD5
A0A1W4X9H0 A0A0C9RYU6 D6X392 A0A1Y1MD29 J3JXX8 N6U031 A0A146LTP3 A0A0A9Z8Y3 A0A182ILW1 A0A0K8TBF4 A0A2J7RRA3 A0A1Q3EXL6 A0A182Q1R4 Q16ML1 A0A2M4BHG3 A0A2M4BI58 A0A182F423 W5JSQ5 A0A2S2Q1X8 A0A2M4BHI1 A0A0P4VPA0 A0A084VKL4 A0A182NBD8 A0A1L8DFQ4 A0A1L8DFP2 B0WU55 A0A2M4AG85 K7IVM8 A0A069DV56 A0A224X9U2 A0A232EQM4 A0A2M3Z3M2 A0A182JV57 A0A1B0CNP2 A0A182VU30 A0A182GHK7 A0A023EUD2 A0A1J1IZU4 A0A0V0G315 E0VUA1 A0A1A8GYV7 A0A1S3JXZ1 A0A182YFR3 A0A336MUU1 U5EH15 A0A067QXA9 A0A3B5LSG5 A0A182RLD3 A0A023F0Q3 J9JZJ3 A0A182LLX0 A0A1L8DFN8 M4A6Q4 A0A1S4GZW1 A0A1A8A9N0 A0A1A9YPW0 A0A182LU10 A0A1B0C529 B4GXB4 A0A2J7RRA4 A0A1A8BUZ5 A0A1A9UWV3 A0A1B0G3J9 A0A3Q2CDM5 Q29K22 J9JUB4 A0A182PED6 A0A1A8KEG0 A0A2H8TZD6 A0A182V710 A0A1A9ZW01 A0A0K8TL75 A0A0L0C3Y4 A0A3P8Z3Q6 A0A1A8QLR3 A0A1A8LYB4 A0A154P9K5 A0A1A7YLF6 A0A034V3T8 A0A0M4E7D0 A0A3Q2QID6 A0A146V7Q4 A0A3P9P4G8 A0A3B0JTV3 B4LQQ2 A0A0A1X2P8 Q4SEX9 A0A1S3R6B4 G3NKD5
Pubmed
19121390
23622113
26354079
18362917
19820115
28004739
+ More
22516182 23537049 26823975 25401762 17510324 20920257 23761445 27129103 24438588 20075255 26334808 28648823 26483478 24945155 20566863 25244985 24845553 25474469 20966253 23542700 12364791 17994087 15632085 26369729 26108605 25069045 25348373 25830018 15496914
22516182 23537049 26823975 25401762 17510324 20920257 23761445 27129103 24438588 20075255 26334808 28648823 26483478 24945155 20566863 25244985 24845553 25474469 20966253 23542700 12364791 17994087 15632085 26369729 26108605 25069045 25348373 25830018 15496914
EMBL
BABH01008444
BABH01008445
NWSH01000782
PCG74237.1
ODYU01010792
SOQ56112.1
+ More
GAIX01004254 JAA88306.1 KQ458793 KPJ05048.1 KQ459984 KPJ19051.1 GBYB01013161 JAG82928.1 KQ971372 EFA10824.2 GEZM01037082 JAV82205.1 BT128100 AEE63061.1 APGK01044418 KB741026 KB631924 ENN74900.1 ERL87176.1 GDHC01008533 JAQ10096.1 GBHO01005339 JAG38265.1 GBRD01002929 JAG62892.1 NEVH01000610 PNF43363.1 GFDL01014984 JAV20061.1 AXCN02000626 CH477858 EAT35579.1 GGFJ01003358 MBW52499.1 GGFJ01003357 MBW52498.1 ADMH02000473 ETN66338.1 GGMS01002437 MBY71640.1 GGFJ01003356 MBW52497.1 GDKW01001765 JAI54830.1 ATLV01014228 KE524948 KFB38508.1 GFDF01008801 JAV05283.1 GFDF01008802 JAV05282.1 DS232100 EDS34774.1 GGFK01006317 MBW39638.1 GBGD01000951 JAC87938.1 GFTR01007310 JAW09116.1 NNAY01002745 OXU20641.1 GGFM01002359 MBW23110.1 AJWK01020587 JXUM01064011 KQ562278 KXJ76252.1 GAPW01000578 JAC13020.1 CVRI01000063 CRL04694.1 GECL01003612 JAP02512.1 DS235784 EEB16957.1 HAEB01010924 HAEC01008260 SBQ76398.1 UFQT01001257 SSX29718.1 GANO01003213 JAB56658.1 KK853247 KDR09373.1 GBBI01004158 JAC14554.1 ABLF02028846 GFDF01008815 JAV05269.1 AAAB01008984 HADY01013347 HAEJ01017641 SBP51832.1 AXCM01000227 JXJN01025803 CH479195 EDW27225.1 PNF43364.1 HADZ01007546 HAEA01008532 SBP71487.1 CCAG010005086 CH379061 EAL33356.2 ABLF02028841 HAEE01010004 SBR30054.1 GFXV01007474 MBW19279.1 GDAI01002670 JAI14933.1 JRES01000940 KNC26990.1 HAEI01005754 SBR94700.1 HAEF01009962 SBR49498.1 KQ434835 KZC07908.1 HADW01001945 HADX01008786 SBP31018.1 GAKP01021780 GAKP01021778 GAKP01021776 JAC37174.1 CP012523 ALC40408.1 GCES01072847 JAR13476.1 OUUW01000010 SPP85547.1 CH940649 EDW63436.1 GBXI01008678 JAD05614.1 CAAE01014610 CAG00803.1
GAIX01004254 JAA88306.1 KQ458793 KPJ05048.1 KQ459984 KPJ19051.1 GBYB01013161 JAG82928.1 KQ971372 EFA10824.2 GEZM01037082 JAV82205.1 BT128100 AEE63061.1 APGK01044418 KB741026 KB631924 ENN74900.1 ERL87176.1 GDHC01008533 JAQ10096.1 GBHO01005339 JAG38265.1 GBRD01002929 JAG62892.1 NEVH01000610 PNF43363.1 GFDL01014984 JAV20061.1 AXCN02000626 CH477858 EAT35579.1 GGFJ01003358 MBW52499.1 GGFJ01003357 MBW52498.1 ADMH02000473 ETN66338.1 GGMS01002437 MBY71640.1 GGFJ01003356 MBW52497.1 GDKW01001765 JAI54830.1 ATLV01014228 KE524948 KFB38508.1 GFDF01008801 JAV05283.1 GFDF01008802 JAV05282.1 DS232100 EDS34774.1 GGFK01006317 MBW39638.1 GBGD01000951 JAC87938.1 GFTR01007310 JAW09116.1 NNAY01002745 OXU20641.1 GGFM01002359 MBW23110.1 AJWK01020587 JXUM01064011 KQ562278 KXJ76252.1 GAPW01000578 JAC13020.1 CVRI01000063 CRL04694.1 GECL01003612 JAP02512.1 DS235784 EEB16957.1 HAEB01010924 HAEC01008260 SBQ76398.1 UFQT01001257 SSX29718.1 GANO01003213 JAB56658.1 KK853247 KDR09373.1 GBBI01004158 JAC14554.1 ABLF02028846 GFDF01008815 JAV05269.1 AAAB01008984 HADY01013347 HAEJ01017641 SBP51832.1 AXCM01000227 JXJN01025803 CH479195 EDW27225.1 PNF43364.1 HADZ01007546 HAEA01008532 SBP71487.1 CCAG010005086 CH379061 EAL33356.2 ABLF02028841 HAEE01010004 SBR30054.1 GFXV01007474 MBW19279.1 GDAI01002670 JAI14933.1 JRES01000940 KNC26990.1 HAEI01005754 SBR94700.1 HAEF01009962 SBR49498.1 KQ434835 KZC07908.1 HADW01001945 HADX01008786 SBP31018.1 GAKP01021780 GAKP01021778 GAKP01021776 JAC37174.1 CP012523 ALC40408.1 GCES01072847 JAR13476.1 OUUW01000010 SPP85547.1 CH940649 EDW63436.1 GBXI01008678 JAD05614.1 CAAE01014610 CAG00803.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000192223
UP000007266
+ More
UP000019118 UP000030742 UP000075880 UP000235965 UP000075886 UP000008820 UP000069272 UP000000673 UP000030765 UP000075884 UP000002320 UP000002358 UP000215335 UP000075881 UP000092461 UP000075920 UP000069940 UP000249989 UP000183832 UP000009046 UP000085678 UP000076408 UP000027135 UP000261380 UP000075900 UP000007819 UP000075882 UP000002852 UP000092443 UP000075883 UP000092460 UP000008744 UP000078200 UP000092444 UP000265020 UP000001819 UP000075885 UP000075903 UP000092445 UP000037069 UP000265140 UP000076502 UP000092553 UP000265000 UP000242638 UP000268350 UP000008792 UP000087266 UP000007635
UP000019118 UP000030742 UP000075880 UP000235965 UP000075886 UP000008820 UP000069272 UP000000673 UP000030765 UP000075884 UP000002320 UP000002358 UP000215335 UP000075881 UP000092461 UP000075920 UP000069940 UP000249989 UP000183832 UP000009046 UP000085678 UP000076408 UP000027135 UP000261380 UP000075900 UP000007819 UP000075882 UP000002852 UP000092443 UP000075883 UP000092460 UP000008744 UP000078200 UP000092444 UP000265020 UP000001819 UP000075885 UP000075903 UP000092445 UP000037069 UP000265140 UP000076502 UP000092553 UP000265000 UP000242638 UP000268350 UP000008792 UP000087266 UP000007635
PRIDE
Pfam
PF05327 RRN3
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9JAT2
A0A2A4JQC3
A0A2H1WT02
S4P8A1
A0A194QHM7
A0A194RSK7
+ More
A0A1W4X9H0 A0A0C9RYU6 D6X392 A0A1Y1MD29 J3JXX8 N6U031 A0A146LTP3 A0A0A9Z8Y3 A0A182ILW1 A0A0K8TBF4 A0A2J7RRA3 A0A1Q3EXL6 A0A182Q1R4 Q16ML1 A0A2M4BHG3 A0A2M4BI58 A0A182F423 W5JSQ5 A0A2S2Q1X8 A0A2M4BHI1 A0A0P4VPA0 A0A084VKL4 A0A182NBD8 A0A1L8DFQ4 A0A1L8DFP2 B0WU55 A0A2M4AG85 K7IVM8 A0A069DV56 A0A224X9U2 A0A232EQM4 A0A2M3Z3M2 A0A182JV57 A0A1B0CNP2 A0A182VU30 A0A182GHK7 A0A023EUD2 A0A1J1IZU4 A0A0V0G315 E0VUA1 A0A1A8GYV7 A0A1S3JXZ1 A0A182YFR3 A0A336MUU1 U5EH15 A0A067QXA9 A0A3B5LSG5 A0A182RLD3 A0A023F0Q3 J9JZJ3 A0A182LLX0 A0A1L8DFN8 M4A6Q4 A0A1S4GZW1 A0A1A8A9N0 A0A1A9YPW0 A0A182LU10 A0A1B0C529 B4GXB4 A0A2J7RRA4 A0A1A8BUZ5 A0A1A9UWV3 A0A1B0G3J9 A0A3Q2CDM5 Q29K22 J9JUB4 A0A182PED6 A0A1A8KEG0 A0A2H8TZD6 A0A182V710 A0A1A9ZW01 A0A0K8TL75 A0A0L0C3Y4 A0A3P8Z3Q6 A0A1A8QLR3 A0A1A8LYB4 A0A154P9K5 A0A1A7YLF6 A0A034V3T8 A0A0M4E7D0 A0A3Q2QID6 A0A146V7Q4 A0A3P9P4G8 A0A3B0JTV3 B4LQQ2 A0A0A1X2P8 Q4SEX9 A0A1S3R6B4 G3NKD5
A0A1W4X9H0 A0A0C9RYU6 D6X392 A0A1Y1MD29 J3JXX8 N6U031 A0A146LTP3 A0A0A9Z8Y3 A0A182ILW1 A0A0K8TBF4 A0A2J7RRA3 A0A1Q3EXL6 A0A182Q1R4 Q16ML1 A0A2M4BHG3 A0A2M4BI58 A0A182F423 W5JSQ5 A0A2S2Q1X8 A0A2M4BHI1 A0A0P4VPA0 A0A084VKL4 A0A182NBD8 A0A1L8DFQ4 A0A1L8DFP2 B0WU55 A0A2M4AG85 K7IVM8 A0A069DV56 A0A224X9U2 A0A232EQM4 A0A2M3Z3M2 A0A182JV57 A0A1B0CNP2 A0A182VU30 A0A182GHK7 A0A023EUD2 A0A1J1IZU4 A0A0V0G315 E0VUA1 A0A1A8GYV7 A0A1S3JXZ1 A0A182YFR3 A0A336MUU1 U5EH15 A0A067QXA9 A0A3B5LSG5 A0A182RLD3 A0A023F0Q3 J9JZJ3 A0A182LLX0 A0A1L8DFN8 M4A6Q4 A0A1S4GZW1 A0A1A8A9N0 A0A1A9YPW0 A0A182LU10 A0A1B0C529 B4GXB4 A0A2J7RRA4 A0A1A8BUZ5 A0A1A9UWV3 A0A1B0G3J9 A0A3Q2CDM5 Q29K22 J9JUB4 A0A182PED6 A0A1A8KEG0 A0A2H8TZD6 A0A182V710 A0A1A9ZW01 A0A0K8TL75 A0A0L0C3Y4 A0A3P8Z3Q6 A0A1A8QLR3 A0A1A8LYB4 A0A154P9K5 A0A1A7YLF6 A0A034V3T8 A0A0M4E7D0 A0A3Q2QID6 A0A146V7Q4 A0A3P9P4G8 A0A3B0JTV3 B4LQQ2 A0A0A1X2P8 Q4SEX9 A0A1S3R6B4 G3NKD5
PDB
3TJ1
E-value=7.03405e-10,
Score=155
Ontologies
PANTHER
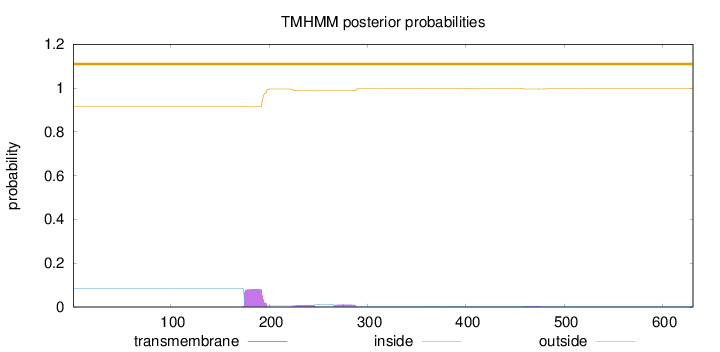
Topology
Length:
631
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.12028
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08450
outside
1 - 631
Population Genetic Test Statistics
Pi
225.355631
Theta
210.948804
Tajima's D
1.031945
CLR
0.124574
CSRT
0.671066446677666
Interpretation
Uncertain
